Circular RNAs (CircRNAs)
Circular RNA (circRNA) is a type of single-stranded RNA covalently closed to form a loop, which occurs by a mechanism known as backsplicing.[1] Due to their circular nature, they have greater stability and are subjected to lesser degradation than linear types of RNA.[2] CircRNAs are mostly produced from protein-coding genes (exons) and multiple circRNAs can arise from a single gene with alternative circularization products, expanding the complexity of post-transcriptional regulation.[3]
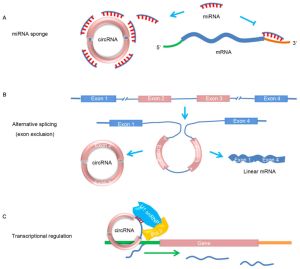
CircRNAs were only discovered relatively recently but are rapidly gaining widespread attention due to their important functions in physiology and their relationship to multiple diseases, such as cancer and age-related diseases. However, how the regulation of circRNAs activity occurs remains largely unknown.
CircRNAs regulate multiple aspects of cell biology.[5][6] In the nucleus, they can lead to epigenetic changes such as histone methylation modifications, alternative splicing alterations or direct interactions with RNA polymerase II, affecting transcription of their gene of origin. In the cytoplasm, they can interact with transcription factors and RNA-binding proteins, induce apoptosis or trap proteins and miRNAs.
Classification of CircRNAs
Circular RNAs fall into four categories based on their genomic locations:
(1) Exonic circular RNAs (ecircRNAs) were found to be processed from exons and are located in the center of RefSeq genes, with potential functions in transcription and gene proliferation (in cytoplasm function as miRNA sponges, interaction with RBP (RNA binding protein), translation);
(2) Circular intronic RNAs (ciRNAs) are found mainly in the nucleus, aiding gene transcription and interacting with RNA polymerase II;
(3) Exon–intron circular RNAs (EIciRNAs) are found in the nucleus and are associated with U1 snRNPs, initiating gene transcription;
(4) tRNA intronic circular RNAs (tricRNAs) are found mainly in the cytoplasm, offering potential in RNA-based applications with relevance to neurodegenerative diseases linked to tRNA processing factors.[7]
The role of circRNAs in aging
CircRNAs shows a stronger correlation with aging than any other type of RNA and has been proposed as a new hallmark of aging.[8]
CircRNAs have been shown to accumulate during aging in the brains of fruit flies,[9] mice[10] and rats.[11] Similarly, C. elegans shows an increase of the vast majority of circRNAs during aging.[12] In human peripheral blood, circRNAs are associated with aging phenotypes and cellular senescence.[13]
In flies, long-lived mutants due to reduced insulin signalling (in a FOXO-dependent manner), show a slower increase in circRNAs accumulation during aging, suggesting that circRNAs accumulation is intrinsically coupled to the rate of aging.[14] The same authors report that circRNAs generated by the sulfateless gene (circSfl) control the lifespan of flies. CircSfl is induced specifically in long-lived mutants and overexpression of circSfl alone is sufficient to extend lifespan in fruit flies.[14]
In worms, two of the most abundant circRNAs accumulating during aging arise from different exons of the crh-1 gene,[12] which corresponds to an ortholog of the cAMP Response Element-Binding Protein (CREB) known to have a role in regulating longevity.[15][16] CRISPR-Cas9 deletion of circ-crh-1 (circular RNA for crh-1 gene), but not crh-1, leads to lifespan extension in worms.[17] This seems to occur mostly in a neuronal tissue-specific manner, as restoration of circ-crh-1 in neurons only leads to a lifespan similar to wild-type. However, it should be noted that the lifespan effects of the removal of circ-crh-1 are rather modest (12%). Nonetheless, selective removal of circRNAs by CRISPR-Cas9, if efficient, provides a powerful tool to study the effect of circRNAs in aging.
For now, it remains unclear whether the age-associated accumulation of circRNAs is detrimental or whether it offers a protective role during aging.
References
- ↑ Li, X., Yang, L., & Chen, L. L. (2018). The biogenesis, functions, and challenges of circular RNAs. Molecular Cell, 71, 428–442.
- ↑ Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., Marzluff, W. F., & Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19, 141–157.
- ↑ Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., & Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell, 159, 134–147.
- ↑ Ren, X. et al. (2017) “Potential functions and implications of circular RNA in gastrointestinal cancer (review),” Oncology Letters [Preprint]. Available at: https://doi.org/10.3892/ol.2017.7118.
- ↑ Braicu, C. et al. (2019) “Comprehensive analysis of circular RNAS in pathological states: Biogenesis, cellular regulation, and therapeutic relevance,” Cellular and Molecular Life Sciences, 76(8), pp. 1559–1577.
- ↑ Ren, X. et al. (2017) “Potential functions and implications of circular RNA in gastrointestinal cancer (review),” Oncology Letters [Preprint]. Available at: https://doi.org/10.3892/ol.2017.7118.
- ↑ Goina, C.A.; Goina, D.M.; Farcas, S.S.; Andreescu, N.I. (2024). The Role of Circular RNA for Early Diagnosis and Improved Management of Patients with Cardiovascular Diseases. Int. J. Mol. Sci., 25(5), 2986; https://doi.org/10.3390/ijms25052986
- ↑ Knupp, D. and Miura, P. (2018) “CircRNA accumulation: A new hallmark of aging?,” Mechanisms of Ageing and Development, 173, pp. 71–79. Available at: https://doi.org/10.1016/j.mad.2018.05.001.
- ↑ Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., Marzluff, W. F., & Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19, 141–157.
- ↑ Gruner, H., Cortes-Lopez, M., Cooper, D. A., Bauer, M., & Miura, P. (2016). CircRNA accumulation in the aging mouse brain. Scientific Reports, 6, 38907.
- ↑ Zhou, T., Xie, X., Li, M., Shi, J., Zhou, J. J., Knox, K. S., Wang, T., Chen, Q., & Gu, W. (2018). Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages. RNA, 24, 1443–1456.
- ↑ 12.0 12.1 Cortes-Lopez, M., Gruner, M. R., Cooper, D. A., Gruner, H. N., Voda, A. I., van der Linden, A. M., & Miura, P. (2018). Global accumulation of circRNAs during aging in Caenorhabditis elegans. BMC Genomics, 19, 8.
- ↑ Haque, S. et al. (2019) “CircRNAs expressed in human peripheral blood are associated with human aging phenotypes, cellular senescence and mouse lifespan,” GeroScience, 42(1), pp. 183–199. Available at: https://doi.org/10.1007/s11357-019-00120-z.
- ↑ 14.0 14.1 Weigelt, C.M. et al. (2020) “An insulin-sensitive circular RNA that regulates lifespan in drosophila,” Molecular Cell, 79(2). Available at: https://doi.org/10.1016/j.molcel.2020.06.011.
- ↑ Chen, Y. C., Chen, H. J., Tseng, W. C., Hsu, J. M., Huang, T. T., Chen, C. H., & Pan, C. L. (2016). A C. elegans thermosensory circuit regulates longevity through crh-1/CREB-dependent flp-6 neuropeptide signaling. Developmental Cell, 39, 209–223.
- ↑ Lakhina, V., Arey, R. N., Kaletsky, R., Kauffman, A., Stein, G., Keyes, W., Xu, D., & Murphy, C. T. (2015). Genome-wide functional analysis of CREB/long-term memory-dependent transcription reveals distinct basal and memory gene expression programs. Neuron, 85, 330–345.
- ↑ Knupp, D. et al. (2022) “Loss of circrnas from the crh‐1 gene extends the mean lifespan in caenorhabditis elegans,” Aging Cell, 21(2).