P53 protein involvement in Longevity
The p53 protein, encoded by TP53 gene, is a specific transcription factor consisting of 393 amino acids with 7 functional domains from N‐terminal to C‐terminal, transactivation domain (TAD)‐1, TAD‐2, proline‐rich domain (PRD), DNA‐binding domain (DBD), hinge domain (HD), oligomerization domain (OD), and C‐terminal regulatory domain (CTR). It is a tumor suppressor that is coded by the most often mutated gene in human cancers.[1] The human gene encoding the p53 protein is called TP53 (italics indicate that this is the name of a gene, not a protein), and includes 11 exons and 10 introns. In humans, this gene is located on chromosome 17 (17p13.1). P53 helps regulate cell growth and repair, and its loss of function is associated with negative outcomes in cancer patients. Activation of p53 is mediated by multiple stress signals, including DNA damage, hypoxia and strong proliferative signals.[1] Depending on the type of stress, p53 can either temporarily halt cell growth and initiate repair or permanently stop cell proliferation.[2]
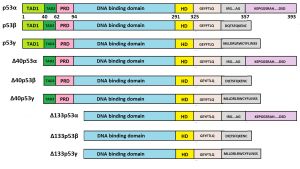
p53 isoforms
Twelve protein isoforms have been shown to be produced through alternative mRNA translation initiation (p53/47) or splicing (full-length p53 or FLp53, p53β, p53γ, Δ40p53α, Δ40p53β, Δ40p53γ, Δ133p53α, Δ133p53β, Δ133p53γ, Δ160p53α, Δ160p53β, Δ160p53γ).[3][4][5]
Hypothesis of Bartas et al.
According to the hypothesis of Bartas et al. specific p53 variations are associated with longevity.[6] In support of the hypothesis, the following arguments are given in particular:
- Bowhead whales had a significantly longer lifespan (about four times longer) compared with other whales. In contrast to other Cetacea, Balaena mysticetus had a unique leucine substitution in the proline-rich region, corresponding to amino acid residue 77 in human p53.
- The olm (Proteus anguinus, Batrachia, Amphibians) has a maximal documented lifespan of 102 years (whereas most amphibian species live for less than 30 years). The p53 protein from this species had additional serine and arginine residues in the core domain (corresponding to an insertion after amino acid L188 in human p53), which had a deleterious effect on p53 functionality.
- Myotis brandtii and Myotis lucifugus are very small bats (max 8 g body weight) and provide a significant exception from Max Kleiber’s law (mouse-to-elephant curve) since their lifespan is extremely long in relation to their small body size. These two species share a unique arrangement in the p53 DNA-binding region, with the insertion of seven amino acid residues in the central DNA-binding region (following amino acid 295 in the human p53 canonical sequence).
The abovementioned analysis and some other facts of long-lived organisms in various animal groups led to conclusion that the amino acid sequence of p53 is associated with organismal lifespan.[6]
TP53 gene copy numbers
Although it is logical to assume that cell divisions increases the chance of mutation accumulation and, consequently, the chance for malignancies, in practice, Peto's paradox is observed: larger animals despite their large body size, a significantly greater number of cells and their long life span not develop cancer more often than much smaller animals with short life span.[7][8] For example, the cancer mortality rate for elephants was found to be less than 5% compared with a cancer mortality rate for humans of 11% to 25%. This may be due to the fact that elephants contain 20 copies of p53, making apoptosis much more likely, whereas humans only possess one copy of p53 in their genetic code, which means that human cells are less likely to trigger apoptosis upon DNA damage, increasing the likelihood of cancer.[9]
For further reading
- Sheekey, E., & Narita, M. (2023). p53 in senescence–it's a marathon, not a sprint. The FEBS journal, 290(5), 1212-1220. https://doi.org/10.1111/febs.16325
- Reinhardt, L. S., Groen, K., Newton, C., & Avery-Kiejda, K. A. (2023). The role of truncated p53 isoforms in the DNA damage response. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 188882. PMID: 36977456 DOI: 10.1016/j.bbcan.2023.188882
- Romani Osbourne, Kelly M. Thayer (2024). Structural and mechanistic diversity in p53-mediated regulation of organismal longevity across taxonomical orders. bioRxiv 2024.08.05.606567; doi: https://doi.org/10.1101/2024.08.05.606567
References
- ↑ 1.0 1.1 Shen, J., Wang, Q., Mao, Y., Gao, W., & Duan, S. (2023). Targeting the p53 signaling pathway in cancers: Molecular mechanisms and clinical studies. MedComm, 4(3), e288. PMID: 37256211 PMCID: PMC10225743 DOI: 10.1002/mco2.288
- ↑ Hafner, A., Bulyk, M. L., Jambhekar, A., & Lahav, G. (2019). The multiple mechanisms that regulate p53 activity and cell fate. Nature reviews Molecular cell biology, 20(4), 199-210. PMID: 30824861 DOI: 10.1038/s41580-019-0110-x
- ↑ Wylie, A., Jones, A. E., Das, S., Lu, W. J., & Abrams, J. M. (2022). Distinct p53 isoforms code for opposing transcriptional outcomes. Developmental cell, 57(15), 1833-1846. PMID: 35820415 PMC9378576 (available on 2023-08-08) DOI: 10.1016/j.devcel.2022.06.015
- ↑ Joruiz, S. M., & Bourdon, J. C. (2016). p53 isoforms: key regulators of the cell fate decision. Cold Spring Harbor perspectives in medicine, 6(8), a026039. PMID: 26801896 PMC4968168 DOI: 10.1101/cshperspect.a026039
- ↑ Anbarasan, T., & Bourdon, J. C. (2019). The emerging landscape of p53 isoforms in physiology, cancer and degenerative diseases. International journal of molecular sciences, 20(24), 6257. PMID: 31835844 PMC6941119 DOI: 10.3390/ijms20246257
- ↑ 6.0 6.1 Bartas, M., Brázda, V., Volná, A., Červeň, J., Pečinka, P., & Zawacka-Pankau, J. E. (2021). The changes in the P53 protein across the animal kingdom point to its involvement in longevity. International Journal of Molecular Sciences, 22(16), 8512. PMID: 34445220 PMCID: PMC8395165 DOI: 10.3390/ijms22168512
- ↑ Tollis, M., Boddy, A. M., & Maley, C. C. (2017). Peto’s Paradox: how has evolution solved the problem of cancer prevention?. BMC biology, 15, 1-5. PMID: 28705195 PMC5513346 DOI: 10.1186/s12915-017-0401-7
- ↑ Perillo, M., Silla, A., Punzo, A., Caliceti, C., Kriete, A., Sell, C., & Lorenzini, A. (2023). Peto’s paradox: nature has used multiple strategies to keep cancer at bay while evolving long lifespans and large body masses. A systematic mini-review. Biomedical Journal, 100654. https://doi.org/10.1016/j.bj.2023.100654
- ↑ Voskarides, K., & Giannopoulou, N. (2023). The role of TP53 in adaptation and evolution. Cells, 12(3), 512. PMID: 36766853 PMCID: PMC9914165 DOI: 10.3390/cells12030512