Autophagy
Autophagy (“auto” meaning “self”, and “phagy” meaning “to eat”) is a catabolic process highly conserved in eukaryotes, in which cytoplasmic components are delivered to the lysosome and degraded. It is a key process for the regulation of cellular energetic equilibrium and plays a housekeeping role by removing cellular waste, damaged organelles, aggregated or misfolded proteins and even intracellular pathogens.[1][2] The process of autophagy is also critical in several developmental processes and in response to nutrient stresses.[3][4] Autophagy promotes cell survival, facilitates bioenergetic homeostasis and can promote cell death.[5]
Types of autophagy
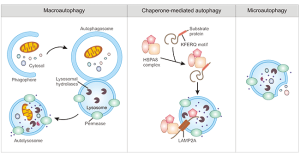
There are three well defined types of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA)[6], with chaperone-assisted selective autophagy (CASA) being recently added.[7][8] Despite differences in their mechanism of degradation and the molecular components they require, all three types of autophagy share in common the delivery of cytoplasmic cargo to the lysosome for proteolytic degradation. The different types of autophagy generally work in a coordinated manner to complement each other.[9]
Macroautophagy
Due to its role in disease, macro-autophagy in particular has been the main focus of research over the past few decades, and often the term “autophagy” is used to refer to the macro-autophagy type of autophagy.[10] Membrane structures in the cytoplasm form autophagosomes, a spherical structure with double-layer membranes
Microautophagy
Microautophagy is a non-selective type of autophagy that involves the delivery of cyoplasmic cargo to the lysosome via autophagic tubes. Microautophagy can be induced under conditions of nitrogen restriction or rapamycin.[9]
Despite microautophagy being generally non-selective, there are three types of selected microautophagy that function in a selective manner: micropexophagy, piecemeal microautophagy of the nucleus (PMN) and micromitophagy.[9]
Chaperone-mediated autophagy (CMA)
Chaperone-mediate autophagy (CMA) is a type of autophagy that specifically engulfs cytosolic proteins, and thus plays an important role in the proteolysis pathway.[11] It is estimated to be responsible for about 30% of protein degradation occurring during caloric restriction, and can also act under conditions of cellular oxidative stress. Similarly to the other types of autophagy, it requires transport of cytosolic cargo to the lysosomes, with the particularity that CMA binds to the LAMP-2A (lysosome-associated membrane protein type 2A) receptor located in the lysosomal membrane.[12] LAMP-2A protein together with a complex of chaperones and co-chaperones are essential for CMA and proteolysis pathways. The expression of LAMP-2A has been shown to decrease with aging, likely due to an age-related increased degradation of LAMP-2A and its decreased ability to insert itself into the lysosomal membrane.[11]
Chaperone-assisted selective autophagy (CASA)
CASA is an autophagy process that selective degrades chaperone-bound proteins in lysosomes.[13] Degradation of primed ubiquitinated chaperones is essential for protein quality control. CASA is critical for muscle maintenance and it is induced in response to resistance exercise in human skeletal muscle.[14] Pathogenic forms of the Huntingtin protein, which may lead to Huntington's disease, are degraded by chaperone-assisted selective autophagy.[15]
Organelle-specific autophagy
There are several types autophagy specific for certain organelles. Removal of damaged or overexpressed organelles is an essential mechanism for cellular quality control and systemic homeostasis.[16] A well studied specific subtype of autophagy is mitophagy (mitochondria). Other types include lysophagy (lysosomes), pexophagy (peroxisomes), reticulophagy (endoplasmic reticulum), ribophagy (ribosomes) and nucleophagy (nucleus).[16]
Mitophagy
The selective degradation of mitochondria through autophagy is known as mitophagy. There are multiple sub-types of mitophagy, such as PINK1/Parkin-dependant, Ubiquitin-independent receptor-mediated and others like piecemeal mitophagy.[17]
Mitophagy declines with age, which leads to the accumulation of damaged mitochondria and the development of a number of age-associated diseases, including Alzheimer's and Parkinson's. Some mitophagy-inducing substances (e.g. resveratrol, metformin and urolithin A) simultaneously activate the biogenesis of new mitochondria, thus promoting health despite aging.[18][19]
Autophagy and longevity
The connections between dysfunctional autophagy with aging and age-related diseases are abundant, and compromised autophagy is considered to be a hallmark of aging.[1][20] Up-regulating autophagy via genetic or pharmacological tools has shown to promote longevity in nematodes, fruit flies and mice.[21][22][23][24][25] On the other hand, in line with these findings, inhibiting selective or non-selective autophagy leads to accelerated aging and functional tissue decline.[1]
Many longevity pathways converge on autophagy genes and can regulate lifespan in animal models such as fruit flies or worms.[26] The process of autophagy is also up-regulated in long-lived C. elegans mutants such as the insulin/insulin-like growth factor (IGF)-1 receptor homolog daf-2 mutant or feeding-defective mutants.[26][27] Furthermore, the 60% lifespan extension of daf-2 loss-of-function mutants is dependent on autophagic genes such as bec-1, lgg-1, atg-7 and atg-12.[28][29][30]
- ↑ 1.0 1.1 1.2 Aman, Y., Schmauck-Medina, T., Hansen, M. et al. Autophagy in healthy aging and disease. Nat Aging1, 634–650 (2021). https://doi.org/10.1038/s43587-021-00098-4
- ↑ Glick, D., Barth, S., & Macleod, K. (2010). Autophagy: cellular and molecular mechanisms. The Journal Of Pathology, 221(1), 3-12. doi: 10.1002/path.2697
- ↑ Maria Fimia, G., Stoykova, A., Romagnoli, A., Giunta, L., Di Bartolomeo, S., & Nardacci, R. et al. (2007). Ambra1 regulates autophagy and development of the nervous system. Nature, 447(7148), 1121-1125. doi: 10.1038/nature05925
- ↑ Qu, X., Zou, Z., Sun, Q., Luby-Phelps, K., Cheng, P., & Hogan, R. et al. (2007). Autophagy Gene-Dependent Clearance of Apoptotic Cells during Embryonic Development. Cell, 128(5), 931-946. doi: 10.1016/j.cell.2006.12.044
- ↑ Das, G., Shravage, B.V. and Baehrecke, E.H. (2012) “Regulation and function of autophagy during cell survival and cell death,” Cold Spring Harbor Perspectives in Biology, 4(6). https://doi.org/10.1101/cshperspect.a008813.
- ↑ Parzych, K., & Klionsky, D. (2014). An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxidants &Amp; Redox Signaling, 20(3), 460-473. doi: 10.1089/ars.2013.5371
- ↑ Ulbricht A, Gehlert S, Leciejewski B, Schiffer T, Bloch W, Höhfeld J. Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy. 2015;11(3):538-46. doi: 10.1080/15548627.2015.1017186. PMID: 25714469; PMCID: PMC4502687.
- ↑ Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, Hoch M, Höhfeld J. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010 Jan 26;20(2):143-8. doi: 10.1016/j.cub.2009.11.022. Epub 2010 Jan 7. PMID: 20060297.
- ↑ 9.0 9.1 9.2 Li, W.-wen, Li, J. and Bao, J.-ku (2011) “Microautophagy: Lesser-known self-eating,” Cellular and Molecular Life Sciences, 69(7), pp. 1125–1136. Available at: https://doi.org/10.1007/s00018-011-0865-5.
- ↑ Ichimiya, T. et al. (2020) “Autophagy and autophagy-related diseases: A Review,” International Journal of Molecular Sciences, 21(23), p. 8974. Available at: https://doi.org/10.3390/ijms21238974.
- ↑ 11.0 11.1 Dice, J.F. (2007) “Chaperone-mediated autophagy,” Autophagy, 3(4), pp. 295–299. Available at: https://doi.org/10.4161/auto.4144.
- ↑ Rout, A.K. et al. (2014) “Structure of transmembrane domain of lysosome-associated membrane protein type 2A (LAMP-2A) reveals key features for substrate specificity in chaperone-mediated autophagy,” Journal of Biological Chemistry, 289(51), pp. 35111–35123. Available at: https://doi.org/10.1074/jbc.m114.609446.
- ↑ Gamerdinger, M. et al. (2009) “Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3,” The EMBO Journal, 28(7), pp. 889–901. Available at: https://doi.org/10.1038/emboj.2009.29.
- ↑ Ulbricht A, Gehlert S, Leciejewski B, Schiffer T, Bloch W, Höhfeld J. Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy. 2015;11(3):538-46. doi: 10.1080/15548627.2015.1017186.
- ↑ Carra S, Seguin SJ, Landry J (January 2008). "HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy". Autophagy. 4 (2): 237–9. doi:10.4161/auto.5407
- ↑ 16.0 16.1 Yao, R.-Q. et al. (2020) “Organelle-specific autophagy in inflammatory diseases: A potential therapeutic target underlying the quality control of multiple organelles,” Autophagy, 17(2), pp. 385–401. https://doi.org/10.1080/15548627.2020.1725377.
- ↑ Bakula D, Scheibye-Knudsen M. MitophAging: Mitophagy in Aging and Disease. Front Cell Dev Biol. 2020 Apr 15;8:239. doi: 10.3389/fcell.2020.00239. PMID: 32373609; PMCID: PMC7179682.
- ↑ Srivastava, V., & Gross, E. (2023). Mitophagy-promoting agents and their ability to promote healthy-aging. Biochemical Society Transactions, BST20221363. PMID: 37650304 DOI: 10.1042/BST20221363
- ↑ Dong, Y., Zhuang, X. X., Wang, Y. T., Tan, J., Feng, D., Li, M., ... & Lu, J. H. (2023). Chemical mitophagy modulators: drug development strategies and novel regulatory mechanisms. Pharmacological Research, 106835. PMID: 37348691 DOI: 10.1016/j.phrs.2023.106835
- ↑ Leidal, A. M., Levine, B. & Debnath, J. Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 20, 1338–1348 (2018).
- ↑ Simonsen, A. et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184 (2008).
- ↑ Pyo, J. O. et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 4, 2300 (2013).
- ↑ Fernandez, A. F. et al. Disruption of the beclin 1–BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140 (2018).
- ↑ Ulgherait, M., Rana, A., Rera, M., Graniel, J. & Walker, D. W. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 8, 1767–1780 (2014).
- ↑ Lapierre, L. R. et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditiselegans. Nat. Commun. 4, 2267 (2013).
- ↑ 26.0 26.1 Tóth, M.L. et al. (2008) “Longevity pathways converge on autophagy genes to regulate life span incaenorhabditis elegans,” Autophagy, 4(3), pp. 330–338. Available at: https://doi.org/10.4161/auto.5618.
- ↑ Mörck, C., Pilon, M. C. elegansfeeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol 6, 39 (2006). https://doi.org/10.1186/1471-213X-6-39
- ↑ Melendez, A. et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 (2003).
- ↑ Hansen, M., Rubinsztein, D. C. & Walker, D. W. Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579–593 (2018).
- ↑ Leidal, A. M., Levine, B. & Debnath, J. Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 20, 1338–1348 (2018).