CHIP phenomenon as a risk factor for atherosclerosis
CHIP (clonal hematopoiesis of indeterminate potential) is an age-related disorder observed in elderly individuals and characterized by the acquisition of somatic mutations in hematopoietic stem cells (HSC) that confer on these cells a selective advantage. As a consequence, instead of the normal polyclonal generation of blood cells, mutation-containing clones expand over time and make up an increasing percentage of the stem cells and their progeny and may include granulocytes, lymphocytes, and monocytes. CHIP is an independent risk factor for myeloid malignancy, cardiovascular disease (CVD), and all-cause mortality. CHIP has also been detected in nonmalignant diseases, such as acquired aplastic anemia (AA), which is an autoimmune disorder. CHIP is rarely found in patients who are younger than 40 years of age, whereas this condition may exist in up to 10% of persons over the age of 70 years.[1][2] CHIP is strongly linked to age acceleration in multiple epigenetic clocks, and the combination of CHIP and epigenetic aging may be used to identify a population at high risk for adverse outcomes and who may be a target for clinical interventions. [3]
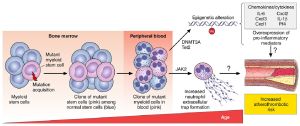
Patients with CHIP have a higher rate of death from noncancer causes (particularly cardiovascular disease) than do age-matched controls without CHIP.[5] CHIP and its common driver genes, in particular, appear to confer an increased risk of coronary heart disease among older persons. [6][7] Four of the most commonly mutated genes in CHIP (DNMT3A, TET2, ASXL1, and JAK2) were each individually associated with coronary heart disease, with the JAK2 V617F mutation carrying 12 times the risk.[6][8][9]
Clonal hematopoiesis in agong
A series of large-scale population-based studies have found that clonal hematopoiesis, driving HSCs dysfunction, increases with age, and the proportion of accumulated mutations, which likely confer a survival advantage in older people reaches one third.[10] [11]
CHIP-associated production of inflammatory cytokines
Among the many risk factors age emerges as the strongest and an independent predictor of cardiovascular disease. The effect of hematopoietic cells on atherosclerosis pathogenesis has long been recognized. The acquisition of genetic mutations in cells, including hematopoietic stem cells, is a feature of aging. Inflammation plays a pivotal role in the pathogenesis of CHIP and its related diseases. Myeloid cells, especially macrophages, derived from HSCs with CHIP-associated mutations exhibit aberrant production of inflammatory cytokines, which gives them a fitness advantage resulting in the propagation of CHIP clones and development of CVD. Genetic IL-6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis
Key genes that have been implicated in CHIP
The most common mutated genes include epigenetic regulators (DNMT3A, TET2, ASXL1), RNA splicing factors (SF3B1, SRSF2, U2AF1, PRPF8), DNA repair and cell cycle regulatory genes (PPM1D, TP53).[12] The hematopoietic function of adults under 65 years of age is polyclonal and has high clonal diversity, while coming forth a sudden and general loss of clonal diversity after the age of 75. Rapidly decreasing clonal diversity is a universal feature of haematopoiesis in aged humans, underpinned by pervasive positive selection acting on many more genes than currently identified.[13] Clonal expansion with abrupt collapse of stem cell diversity may be a feature of ageing that generalizes beyond human blood. Many human organ systems studied to date, including skin, bronchus, endometrium, and oesophagus, show age-related expansions of clones with driver mutations. With such ubiquity of driver mutations—selected purely for their competitive advantage within the stem cell compartment—and with the wholesale rewiring of cellular pathways they induce, it is feasible that they may contribute to phenotypes of human ageing beyond the risk of cancer. Mutations of loss-of-function methylcytosine dioxygenase 2 (TET2), DNA (cytosine-5)-methyltransferase 3A (DNTM3A), polycomb chromatin-binding protein (ASXL1), and gain-of-function JAK2 transcriptional regulator genes result in positive selection and expansion of clones of hematopoietic cells without malignancy. TET2 or JAK2 clonal mutation increases IL-6 and IL-1β production in myeloid cells supporting accelerated atherosclerosis.
| Major Genes Harboring Somatic Mutations Associated With CHIP[14] | Description of Gene Product | |
|---|---|---|
| DNMT3A | Genome-wide DNA methyltransferase causing DNA demethylation | |
| TET2 | DNA binding protein that mediates conversion of methylcytosine to 5-hydroxymethylcytosine and causes DNA demethylation | |
| ASXL1 | Chromatin binding protein that regulates polycomb-mediated transcriptional repression | |
| TP53 | Transcription factor that governs apoptosis, DNA repair, senescence, and cell stress–induced responses | |
| JAK2 | Protein tyrosine kinase involved in broad cell growth, differentiation, and developmental programs |
Janus-associated kinase 2 (JAK2)
Janus-associated kinase 2 (JAK2) is one member of a family of four cytoplasmic tyrosine kinases that also includes JAK1, JAK3 and Tyk2. The JAK enzymes are required for signaling by cytokine and growth factor receptors that lack intrinsic kinase activity. There appear to be some overlapping roles for JAK family members, as most signaling pathways involve more than one JAK, with the exception of some growth factors such as erythropoietin and thrombopoietin, which only utilize JAK2.[16]
While JAK2 inhibitors may prove to be effective in controlling hyperproliferation of hematopoietic cells in a wide range of diseases, they may not be able to eliminate mutant clones.[17]
Key genes that have been implicated in atherosclerosis
Six hub genes for atherosclerosis were identified, which effectively distinguished between normal and atherosclerosis samples, among which ADAP2, CSF3R, FABP5, ITGAX, and SPP1 were highly expressed in atherosclerosis, whereas MYOC expression was downregulated.[21]
ADAP2 encodes ArfGAP with the double PH domain 2 protein, which plays an important role in cardiac development.
CSF3R is essential for granulocyte maturation and is primarily involved in the regulation of inflammation.
FABP5 plays a role in the transportation, regulation, and metabolism of lipids. FABP5 is a sensitive marker for lipid-rich macrophages on the luminal side of atherosclerosis lesions.
ITGAX is involved in early atherosclerosis lesion formation and is highly expressed in atherosclerosis tissues.
SPP1 exhibits high expression in endothelial cells, macrophages, and vascular smooth muscle cells present in atherosclerotic plaques. MYOC and its secreted glycoproteins may influence atherosclerosis by regulating processes such as cell adhesion, cytoskeleton, and migration
References
- ↑ Steensma, D. P., Bejar, R., Jaiswal, S., Lindsley, R. C., Sekeres, M. A., Hasserjian, R. P., & Ebert, B. L. (2015). Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood, The Journal of the American Society of Hematology, 126(1), 9-16. PMID: 25931582 PMC4624443 DOI: 10.1182/blood-2015-03-631747
- ↑ Natarajan, P., Jaiswal, S., & Kathiresan, S. (2018). Clonal hematopoiesis: somatic mutations in blood cells and atherosclerosis. Circulation: Genomic and Precision Medicine, 11(7), e001926. PMID: 29987111 PMC6082163 DOI: 10.1161/CIRCGEN.118.001926
- ↑ Nachun, D., Lu, A. T., Bick, A. G., Natarajan, P., Weinstock, J., Szeto, M. D., ... & NHLBI Trans‐Omics for Precision Medicine (TOPMed) Consortium. (2021). Clonal hematopoiesis associated with epigenetic aging and clinical outcomes. Aging Cell, 20(6), e13366. PMID: 34050697 PMC8208788 DOI: 10.1111/acel.13366
- ↑ According to Libby, P., & Ebert, B. L. (2018). CHIP (clonal hematopoiesis of indeterminate potential) potent and newly recognized contributor to cardiovascular risk. Circulation, 138(7), 666-668. PMID: 30359133 PMC6277144 DOI: 10.1161/CIRCULATIONAHA.118.034392
- ↑ Jaiswal, S., Fontanillas, P., Flannick, J., Manning, A., Grauman, P. V., Mar, B. G., ... & Ebert, B. L. (2014). Age-related clonal hematopoiesis associated with adverse outcomes. New England Journal of Medicine, 371(26), 2488-2498. PMID: 25426837 PMC4306669 DOI: 10.1056/NEJMoa1408617
- ↑ 6.0 6.1 Jaiswal, S., Natarajan, P., Silver, A. J., Gibson, C. J., Bick, A. G., Shvartz, E., ... & Ebert, B. L. (2017). Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. New England Journal of Medicine, 377(2), 111-121. PMID: 28636844 PMC: 6717509 DOI: 10.1056/NEJMoa1701719
- ↑ Keaney Jr, J. F. (2017). CHIP-ping away at atherosclerosis. New England Journal of Medicine, 377(2), 184-185. PMID: 28636833 DOI: 10.1056/NEJMe1706173
- ↑ Libby, P., Jaiswal, S., Lin, A. E., & Ebert, B. L. (2019). CHIPping away at the pathogenesis of heart failure. JAMA cardiology, 4(1), 5-6.
- ↑ Dorsheimer, L., Assmus, B., Rasper, T., Ortmann, C. A., Ecke, A., Abou-El-Ardat, K., ... & Rieger, M. A. (2019). Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA cardiology, 4(1), 25-33. PMID: 30566180 PMC6439691 DOI: 10.1001/jamacardio.2018.3965
- ↑ Kanagal-Shamanna, R., Beck, D. B., & Calvo, K. R. (2023). Clonal hematopoiesis, inflammation, and hematologic malignancy. Annual Review of Pathology: Mechanisms of Disease, 19. PMID: 37832948 DOI: 10.1146/annurev-pathmechdis-051222-122724
- ↑ Belotserkovskaya, E., Golotin, V., Uyanik, B., & Demidov, O. N. (2023). Clonal haematopoiesis-a novel entity that modifies pathological processes in elderly. Cell Death Discovery, 9(1), 345. PMID: 37726289 PMC10509183 DOI: 10.1038/s41420-023-01590-z
- ↑ Aging Biomarker Consortium., Bao, H., Cao, J. et al. (2023). Biomarkers of aging. Sci. China Life Sci. https://doi.org/10.1007/s11427-023-2305-0
- ↑ Mitchell, E., Spencer Chapman, M., Williams, N., Dawson, K. J., Mende, N., Calderbank, E. F., ... & Campbell, P. J. (2022). Clonal dynamics of haematopoiesis across the human lifespan. Nature, 606(7913), 343-350. PMID: 35650442 PMC9177428 DOI: 10.1038/s41586-022-04786-y
- ↑ Khetarpal, S. A., Qamar, A., Bick, A. G., Fuster, J. J., Kathiresan, S., Jaiswal, S., & Natarajan, P. (2019). Clonal hematopoiesis of indeterminate potential reshapes age-related CVD: JACC review topic of the week. Journal of the American College of Cardiology, 74(4), 578-586.PMID: 31345433 PMC6662618 DOI: 10.1016/j.jacc.2019.05.045
- ↑ Buscarlet, M., Provost, S., Zada, Y. F., Barhdadi, A., Bourgoin, V., Lépine, G., ... & Busque, L. (2017). DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood, The Journal of the American Society of Hematology, 130(6), 753-762. PMID: 28655780 DOI: 10.1182/blood-2017-04-777029
- ↑ Vainchenker, W., Dusa, A., & Constantinescu, S. N. (2008, August). JAKs in pathology: role of Janus kinases in hematopoietic malignancies and immunodeficiencies. In Seminars in cell & developmental biology (Vol. 19, No. 4, pp. 385-393). Academic Press. PMID: 18682296 DOI: 10.1016/j.semcdb.2008.07.002
- ↑ Misaka, T., Kimishima, Y., Yokokawa, T., Ikeda, K., & Takeishi, Y. (2023). Clonal hematopoiesis and cardiovascular diseases: role of JAK2V617F. Journal of Cardiology, 81(1), 3-9. PMID: 35165011 DOI: 10.1016/j.jjcc.2022.02.001
- ↑ Zhang, J. Y., Sun, J. F., Nie, P., Herdewijn, P., & Wang, Y. T. (2023). Synthesis and clinical application of small-molecule inhibitors of Janus kinase. European Journal of Medicinal Chemistry, 115848. PMID: 37793326 DOI: 10.1016/j.ejmech.2023.115848
- ↑ Roskoski Jr, R. (2022). Janus kinase (JAK) inhibitors in the treatment of neoplastic and inflammatory disorders. Pharmacological Research, 106362. PMID: 35878738 DOI: 10.1016/j.phrs.2022.106362
- ↑ Verstovsek, S. (2009). Therapeutic potential of JAK2 inhibitors. ASH Education Program Book, 2009(1), 636-642. PMID: 20008249 PMC5166576 DOI: 10.1182/asheducation-2009.1.636
- ↑ Wu, X., Pan, X., Zhou, Y. et al. (2024). Identification of key genes for atherosclerosis in different arterial beds. Sci Rep 14, 6543 https://doi.org/10.1038/s41598-024-55575-8