Epigenetic reprogramming
Epigenetic reprogramming generally refers to a significant global remodeling of epigenetic features of DNA. Historically, this term has referred to changes occurring in the context of early organismal development. It has also been referenced in relation to cancer and other disease states.[1][2]
Within the context of longevity research, the term has more specifically been used to refer to a new strategy for cellular rejuvenation.[3][4][5] This article focuses on a narrower definition of epigenetic reprogramming known as reprogramming-induced rejuvenation (RIR), which aims to induce in vivo cellular rejuvenation to continually rewind the clock of aging as a way to prevent, treat, or reverse the diseases of aging.[5][6][7]
Interventions so far indicate that epigenetic reprogramming is capable of rejuvenating animal tissues as seen by both aging clocks and functional markers. Importantly, there is first evidence that epigenetic reprogramming in wild type animals is able to extend lifespan. Macip et al. (2023)[8] observed a significant effect of partial reprogramming on late stage median lifespan when applied to aged wild type mice. Using an adeno-associated virus 9 (AAV9) vector to systemically deliver OSK to 124-week-old mice, the group showed that IVPR could extend the remaining median lifespan of treated mice by 109%.[8][9]
Epigenetics
Epigenetics refers to the study of heritable yet modifiable features or marks on the genome which contribute to gene expression. These features impact the chromatin structure without any change in the nucleotide sequence of DNA and function to regulate how genes are transcribed into proteins.[1][10]
The forms of youthful information storage have been reported to include DNA modifications, histone modifications, RNA modifications, noncoding (nc)RNAs, DNA–RNA hybrids such as R-loops, and protein–DNA interactions.[11][12]
The most prominent and well-studied of these epigenetic features is DNA methylation.[13][14] Measurements of DNA methylation patterns have also been used to form the basis for epigenetic aging clocks, as a potential measure of biological age across the mammalian kingdom.[15] Other epigenetic changes include histone modifications and non-coding RNA (ncRNA) mediated gene silencing.[10]
Epigenetic reprogramming as a rejuvenation strategy
Reprogramming induced rejuvenation
Applying reprogramming induced rejuvenation (RIR) in naturally aged mice has recently been shown to improve memory and cognition, promote muscle regeneration, and restore vision.[4][16][17] Analyses using epigenetic aging clocks, which measure some aspects of biological age, show that cellular reprogramming resets epigenetic age.[18][9]
It has been envisioned by some scientists as a therapy that could be used periodically, perhaps every few decades, to continually reverse aging in humans.[19]
Information theory of aging
David Sinclair of Harvard Medical School has theorized that progressive loss of youthful epigenetic information not only is associated with aging, but is a key driver of aging.[4] Sinclair uses the analogy of a compact disc with scratches that can be polished off to recover function; similarly, it is hypothesized that the epigenome can be recovered to its youthful state.[4][19] Drawing from Claude Shannon's information theory, Sinclair speculates that cells retain an original copy of information as backup as a countermeasure against information loss or noise.[20] Storing such information could involve DNA-binding proteins, covalent DNA changes, RNA-guided chromating modifying factors, and RNA-DNA hybrids.[4] Further support of this theory placing epigenetic changes at the center of aging requires more empirical evidence, particularly in considering the novel nature of reprogramming research.
Epigenetic reprogramming contexts
In development
Two major reprogramming events are known to occur in early development of mammals, in early embryos and primordial germ cells. During these events, methylation patterns are reprogrammed genome wide.[21][22]
In cancer
All cancer cells are known to have epigenetic changes.[23] These changes lead to induced pluripotency, the transformation of differentiated cells into so-called cancer stem cells.[23][24] Transcription factors initiate DNA transcription and work synergistically with epigenomic regulators in the promotion of cancer.[25] Modification of aberrant epigenetic marks has been proposed as a strategy for targeting cancer, utilizing solely epi-drugs or in combination with chemotherapy or immunotherapy.[26] One example of this is epigenome editing (e-GE) to hypermethylate (suppress) oncogenes or hypomethylate (upregulate) tumor suppressor genes.[27] Additionally, e-GE may allow for improved responses or lower effective doses when used along side existing treatments.[27]
In disease states
Epigenetic variability has been noted as a driving force for a multitude of diseases.[2] It is particularly a feature of age-related diseases, including cardiovascular disease, Type 2 diabetes and dementia.[28]
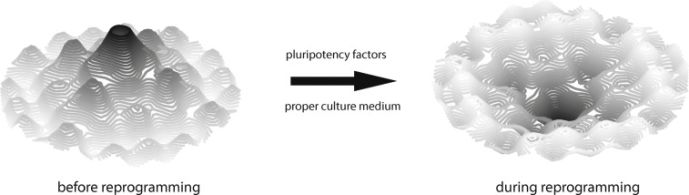
The epigenetic landscape
In 1957, Conrad Waddington, often referred to as the father of epigenetics, purposed an analogy of a landscape to describe the plasticity occurring during development in response to environmental factors. In the landscape, a marble starts atop an elevation to represent a point of stochastic state, or if referring to cell biology, a pluripotent state. As the marble rolls down the landscape, it encounters different troughs and valleys representing separate paths it may take to its final phenotype or differentiated state.[29][30] This landscape analogy can be useful for visualizing epigenetic transition during reprogramming.[30] Using such imagery, reprogramming reshapes the landscape so that the peaks and troughs invert.
Epigenetic reprogramming strategies for rejuvenation
Because epigenome editing does not involve modification of the genome itself, it is currently considered to have a lower impact on germ cells than genome editing. Thus, epigenome editing has the potential to overcome important scientific and ethical issues of concern with genome editing. Various efforts are underway to successfully apply epigenome editing in vivo, such as improving target specificity, enzymatic activity, and drug delivery for the development of reliable therapeutics.[31]
Yamanaka factors
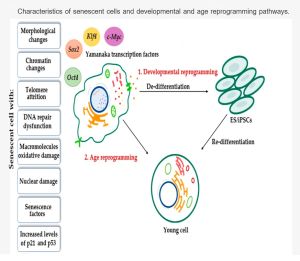
Epigenetic reprogramming for rejuvenation is based on work that earnt Shinya Yamanaka the 2012 Nobel Prize in Medicine. Yamanaka showed that it was possible to reprogram adult body cells back into biologically immortal pluripotent stem cells capable of turning into any cell type.[32][33] These reprogrammed cells show profound reversion of cellular identity, accompanied by a ‘youthful’ embryonic epigenetic state. This is achieved by activating only four transcription factors - the Yamanaka factors - OCT4, SOX2, KLF4, and MYC (or OSKM).[32][33] These transcription factors cause epigenetic changes that alter gene expression, instead of making changes to DNA itself.[32][33] While Yamanaka reprogramming into induced pluripotent stem cells results in a rejuvenated phenotype, it is not a practical solution for targeting aging.[5] This is because it results in a loss of cellular identity, and critically, the acquired ability to self-renew inevitably leads to teratomas in mouse models.[34][5]
Partial reprogramming using Yamanaka factors
Scientists in the aging biology field have since expanded upon this work to show that a partial version of this epigenetic reprogramming technique may reverse multiple aspects of aging.[34] Work from Juan Carlos Izpisua Belmonte's lab showed that it is possible to modify the reprogramming technique and achieve 'youthful' rejuvenation, without resetting a cell with a defined identity into a stem cell. This is known as partial reprogramming, and commonly referred to as epigenetic reprogramming. Epigenetic aging clocks, which measure the methylation status of various tissues and predict biological age, have been observed to be reset with partial cellular reprogramming.[36]
The Belmonte lab showed that aging could be slowed via cyclic induction of reprogramming by extending lifespan in a mouse model of Hutchinson-Gilford progeria syndrome, a form of accelerated aging. Normally aged 12-month-old mice treated with the same strategy showed a recovery of pancreatic islet function and improvements in muscle function following acute injury. Whether partial reprogramming can reliably slow aging in normally aged mice remains an unanswered question.
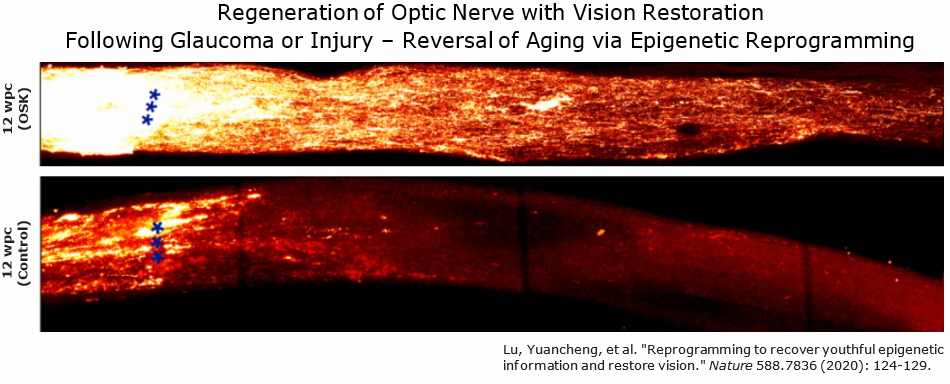
Systemic approaches to epigenetic reprogramming may not always be feasible due to greater complexity and reduced control over variables that may affect transcription factor expression. It has since been investigated more locally, such as within the eye, as opposed to systemically throughout the body. Regeneration of the optic nerve and visual recovery was demonstrated in a mouse model of glaucoma and natural aging, by investigating the effects of in vivo reprogramming on rejuvenation.[4] The strategy used by the David Sinclair lab to achieve partial reprogramming without inducing teratomas involved omission of the MYC oncogene from the original Yamanaka OSKM protocol, known as OSK. Instead, OSK expression was controlled and induced via doxycycline, delivered via a Tet-ON AAV9 system.[4] While no teratomas were observed using this strategy, in the absence of more data, some level of theoretical risk remains because KLF4 and SOX2 are linked to cancer-related pathways.[37]
It is notable that for the first time, reprogramming with OSK enables recovery of anatomy and vision after retinal damage and visual decline has already occurred in mice. This contrasts with various other previous approaches which have typically required treatment in early stages of disease, in order to show reduced progression. Epigenetic rejuvenation has the potential to not only address early glaucoma, but also an unmet need in late-stage glaucoma patients who have already lost retinal ganglion cells and functional vision.
Studies have demonstrated that partial reprogramming using the Yamanaka factors (or a subset; OCT4, SOX2, and KLF4; OSK) not only can reverse age-related changes in vitro and in vivo, but are also capable of extending the lifespan, for example, of aged wild type mice. Systemically delivered AAVs, encoding an inducible OSK system, in 124-week-old mice extends the median remaining lifespan by 109% over wild-type controls and enhances several health parameters.[8][9]
Multipotent transcription factors
Multipotent cells, which are cells that are not fully reprogrammed to iPSCs yet still partially undifferentiated, have been investigated for reprogramming. This RIR strategy may mitigate the risk of oncogenesis.[3]
Researchers from the Alphabet Inc. (Google) subsidiary, Calico, analyzed different combinations of Yamanaka factors on different cell types, finding that they restored youthful gene expression, but also affected cell identity. Most importantly, they found any combination of factors had the potential to cause pluripotency. This suggests that even transient reprogramming can be oncogenic, with more transcription factors conferring higher risk. However, the suppression of cell identity and restoration of youthful gene expression may perhaps be decoupled, so that there may be as yet discovered methods which restore a youthful phenotype with low risk for pluripotency.[3]
Reprogramming with small molecules
The safety of solely introducing exogenous transcription factors in Yamanaka factor reprogramming is controversial due to potential cancer risk from gene mutations or insertions.[5][38] Strategies mentioned previously involve refining the original iPSC reprogramming transcription factors, while the use of small molecules is an alternate method for epigenetic reprogramming.[5][38][39] This strategy is also being pursued as it may have the potential to improve the safety and efficiency of exogenous transcription factor reprogramming.[38] For example, it was recently shown that partial epigenetic reprogramming using a combination of small molecules was able to improve liver regeneration and function in a mouse model of acute liver injury.[40] A variety of chemical cocktails are known to be capable of rejuvenating cells and reversing transcriptomic age to a similar extent as OSK overexpression.[41][42]
Vitamin C
Many mammals, synthesize ascorbic acid in the liver using the enzyme gulonolactone oxidase, whereas humans are dependent on dietary intake of vitamin C. Sufficient intake of vitamin C could play a role in the prevention of tumors. It is assumed that vitamin C alleviates the methylation defect observed at enhancer regions.[43] Vitamin C can enhance epigenetic reprogramming of fibroblasts to induced pluripotent stem cells (iPSCs), and hinder the aberrant self-renewal of hematopoietic stem cells (HSCs) through its ability to enhance the activity of either Jumonji C (JmjC) domain-containing histone demethylases or ten-eleven translocation (TET) DNA hydroxylases.[44][45]
Methods for introducing reprogramming factors
As RIR strategies require the introduction of exogenous reprogramming genes in target cells, delivery is critical for appropriate gene expression at both the desired location and to the required extent.
Viral vectors
Reprogramming factors may be delivered via various viral delivery systems, such as an adeno-associated viral vector (AAV). As applied in a mouse model of glaucoma, the specific engineered viral vector, Tet-Off AAV2, uses tetracycline class drugs as an on-off switch to induce transcription for a controlled duration.[4]
Messenger RNA (mRNA)
In addition to using AAV technology to introduce transcription factors, mRNA technology can also be employed. Turn Biotechnology is an example of a company currently investigating mRNA-based technology for epigenetic reprograming.[46]
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9-based genomic editing
CRISPR/Cas9-based methods have also been used to induce reprogramming through modulating methylation at specific CpG sites and eliciting gene expression of specific genes of epigenetic reprogramming regulators (such as Yamanaka factors).[47] This methodology has demonstrated the ability to deliver the Yamanaka factors to produce iPSCs.[47]
Pharmaceutical/nutritional interventions
There is hope that it may be possible to induce similar rejuvenating epigenetic reprogramming via pharmaceuticals or supplements, however research in this area is presently in its infancy. This approach is advantageous as it avoids direct gene editing and use of the Yamanaka factors, resulting in better safety and access. It is speculated though, that the rejuvenative effect may not be as robust and there is evidence that nutritional requirements for longevity may vary within a population.[48][49]
Limitations/challenges
Reprogramming to iPSC
As mentioned previously, a major limitation of RIR is the potential for dedifferentiation into a pluripotent state that results in the formation of tumors. More prudent approaches such as partial reprogramming may mitigate this risk, as doing so may induce rejuvenation without dedifferentiation. However, research in this area in vivo is still limited and has thus far been limited only to mouse models. It has been theorized that it may be possible to separate pluripotency from rejuvenation, which may have important implications for improving safety.[3][50]
Viral vector risks
Retroviral vectors commonly utilized in laboratory studies risk insertional mutagenesis, residual expression/re-activation of reprogramming factors, and retrotransposon activation.[5] This could be averted through transient transfection, non-integrating viral vectors, RNA transfection and chemical reprogramming via small molecules.[5] Viral vectors also present challenges related to immunogenicity, biodistribution, and transduction efficiency.[51]
Delivery
Whole organismal delivery of RIR presents multiple hurdles in complex organisms. First, dispersion to the entire organism, or biodistribution, is a challenge as the treatment may accumulate variably, more in certain tissues and less or none in others. A second challenge is that the cocktail of factors and/or molecules may need to be tailored to the specific cell type. Essentially, the therapy needs to be distributed across the organism in the correct amounts with mixtures specific to the each cell type.
Resistance to in vivo reprogramming
In addition to delivery challenges there is variation in susceptibility of cells to epigenetic reprogramming. Susceptibility varies by cell type.[3] Also, senescent cells are known to be resistant to reprogramming, though this may be amenable to adjusting the cocktail of factors or possibly through multiple rounds of reprogramming.[52] or depletion of p16High senescence.[53] Senescent human fibroblast cells have been successfully reprogrammed to pluripotent cells utilizing the canonical Yamanaka factors, in addition to NANOG and LIN28.[52]
However, not only are senescent cells resistant to Yamanaka factor reprogramming, reprogramming itself may induce cellular senescence in other cells. Additionally, senescent cell activity and/or tissue damage may be a prerequisite for successful in vivo reprogramming.[54][55][56] Using RIR and senescent cell elimination in combination has been proposed as way to optimize tissue regeneration and rejuvenation.[57] Compounds that mimic cellular senescence, like the drug Palbociclib, have been shown to improve reprogramming efficiency.[55] The immune system of the aged somewhat resembles that of a newborn: compromised lymphocyte responses, reduced activity of macrophages and neutrophils, decreased natural killer (NK) cell killing, and reduced antigen presentation by dendritic cells.[58] Consequently, the efficacy of in vivo partial reprogramming should significantly increase in the elderly due to the depletion of NK cells, which act as an extrinsic barrier for in vivo reprogramming.[59]
Age-reversal-age- extension (Arae) paradox
Several lifespan-extending interventions, particularly those that work on metabolic pathways (e.g. mTOR, AMPK), promote genomic stability. Genomic stability creates deeper valleys in the epigenetic landscape while reprogramming flattens the landscape so that the marble is more free to move about. The ideal reprogramming method would carve out a valley on the epigenetic landscape leading straight to the desired state.[30]
Companies pursuing epigenetic reprogramming
AgeX Theraputics
Mission Statement: "The mission of AgeX is to develop and commercialize novel therapeutics targeting biological aging based on an emerging understanding of the ‘clockwork mechanisms’ of human aging. We plan to apply these technologies in the practice of human medicine to extend human health and life spans."
The company is developing multiple platforms for potential first-in-class therapeutic cell therapies, small-molecule drugs and medical devices. The company states their platform called induced Tissue Regeneration (iTR™) uses a proprietary formulation labeled AGEX-iTR1547, which has demonstrated initial capability of reducing the expression of the marker gene related to lost regenerative potential and is referred to by the company as partial reprogramming.[60]
Altos Labs
First reported by MIT Technology Review on September 04, 2021.[61] Reportedly funded by Jeff Bezos and Yuri Milner to pursue lifespan extension via "biological reprogramming technology". The company has recruited multiple leading aging biology scientists from academia, including Juan Carlos Izpisúa Belmonte, Manuel Serrano and Steve Horvath, among dozens of other scientists from geroscience and adjacent fields.
Nobel laureate Shinya Yamanaka is an unpaid senior scientist and chair of the company’s scientific advisory board, along with other Nobel awardees Jennifer Doudna, Frances Arnold, and David Baltimore.[62]
Altos chief scientist and founder is Richard Klausner, a previous director of the US National Cancer Institute, and former Executive Director of the Gates Foundation.[63] Klausner is also founder of GRAIL, a cancer detection company; co-founder of Juno Therapeutics, a cancer immunotherapy company; and previously the Senior Vice President of Illumina.[64]
Calico Labs
Mission Statement: "Calico (Calico Life Sciences LLC) is an Alphabet-founded research and development company whose mission is to harness advanced technologies and model systems to increase our understanding of the biology that controls human aging. Calico will use that knowledge to devise interventions that enable people to lead longer and healthier lives."
Posted preprint research on May 23, 2021 mapped trajectories of partial reprogramming in multiple cell types using single cell genomics. The research also explores partial multipotent reprogramming in myogenic cells.[3][65]
Life Biosciences
The proprietary gene therapy platform aims to express OSK, comprising three of the four Yamanaka factors, to reprogram the aged epigenome to a younger state. The gene therapy has been shown to restore vision in a mouse glaucoma model and in mice with aging-related vision loss.[66]
Shift Bioscience
Using machine learning to evaluate public and proprietary gene expression data from cell reprogramming studies to identify the contributions that different genes make to the rejuvenation process, with the goal of safely resetting cells and tissues back to a youthful state.[50]
Turn Biotechnologies
The proprietary epigenetic reprogramming of age (ERA™) Platform uses mRNA to deliver transcription factors to the epigenome. The time, duration and dosage of transcription factors are controlled to optimize the mRNA cocktail for each indication and tailored specifically for tissue type.[46]
References
- ↑ 1.0 1.1 Moosavi, A., & Ardekani, A. M. (2016). Role of epigenetics in biology and human diseases. In Iranian Biomedical Journal (Vol. 20, Issue 5, pp. 246–258). Pasteur Institute of Iran. https://doi.org/10.22045/ibj.2016.01
- ↑ 2.0 2.1 Feinberg, A. P. (2018). The key role of epigenetics in human disease prevention and mitigation. New England Journal of Medicine, 378(14), 1323-1334.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Roux, A., Zhang, C., Paw, J., Zavala-Solorio, J., Vijay, T., Kolumam, G., Kenyon, C., & Kimmel, J. C. (2021). Partial reprogramming restores youthful gene expression through transient suppression of cell identity. BioRxiv, 2021.05.21.444556.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 Lu, Y., Brommer, B., Tian, X., Krishnan, A., Meer, M., Wang, C., Vera, D. L., Zeng, Q., Yu, D., Bonkowski, M. S., Yang, J. H., Zhou, S., Hoffmann, E. M., Karg, M. M., Schultz, M. B., Kane, A. E., Davidsohn, N., Korobkina, E., Chwalek, K., … Sinclair, D. A. (2020). Reprogramming to recover youthful epigenetic information and restore vision. Nature, 588(7836), 124–129. https://doi.org/10.1038/s41586-020-2975-4
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Simpson, D. J., Olova, N. N., & Chandra, T. (2021). Cellular reprogramming and epigenetic rejuvenation. Clinical Epigenetics, 13(1), 1-10.
- ↑ Yu, J., Li, T., & Zhu, J. (2023). Gene Therapy Strategies Targeting Aging-Related Diseases. Aging and disease, 14(2), 398. PMID: 37008065 PMC: 10017145 DOI: 10.14336/AD.2022.00725
- ↑ Yücel, A. D., & Gladyshev, V. N. (2024). The long and winding road of reprogramming-induced rejuvenation. Nature Communications, 15(1), 1941. PMID: 38431638 PMC10908844 DOI: 10.1038/s41467-024-46020-5
- ↑ 8.0 8.1 8.2 Macip, C. C., Hasan, R., Hoznek, V., Kim, J., Metzger IV, L. E., Sethna, S., & Davidsohn, N. (2023). Gene therapy mediated partial reprogramming extends lifespan and reverses age-related changes in aged mice. bioRxiv, 2023-01. Doi: 10.1101/2023.01.04.522507 also: (Feb 2024). Cellular Reprogramming, 26(1). Doi: 10.1089/cell.2023.0072
- ↑ 9.0 9.1 9.2 Paine P.T., Ada Nguyen, Ocampo A. (2023). Partial cellular reprogramming: A deep dive into an emerging rejuvenation technology. https://doi.org/10.1111/acel.14039
- ↑ 10.0 10.1 Genetics, Epigenetic Mechanism - StatPearls - NCBI Bookshelf. (n.d.). Retrieved September 8, 2021, from https://www.ncbi.nlm.nih.gov/books/NBK532999/?report=classic
- ↑ Aging Biomarker Consortium, Bao, H., Cao, J., Chen, M., Chen, M., Chen, W., ... & Liu, G. H. (2023). Biomarkers of aging. Science China Life Sciences, 66(5), 893-1066. PMID: 37076725 PMC10115486 DOI: 10.1007/s11427-023-2305-0
- ↑ Wu, Z., Zhang, W., Qu, J., & Liu, G. H. (2024). Emerging epigenetic insights into aging mechanisms and interventions. Trends in Pharmacological Sciences. 45(2), 157-172 PMID: 38216430 DOI: 10.1016/j.tips.2023.12.002
- ↑ Reik, W., Dean, W., & Walter, J. (2001). Epigenetic reprogramming in mammalian development. In Science (Vol. 293, Issue 5532, pp. 1089–1093). American Association for the Advancement of Science. https://doi.org/10.1126/science.1063443
- ↑ Li, Y. (2021). Modern epigenetics methods in biological research. Methods, 187, 104–113. https://doi.org/10.1016/J.YMETH.2020.06.022
- ↑ Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14(10). https://doi.org/10.1186/gb-2013-14-10-r115
- ↑ Wang, C., Ros, R. R., Martinez-Redondo, P., Ma, Z., Shi, L., Xue, Y., ... & Belmonte, J. C. I. (2021). In vivo partial reprogramming of myofibers promotes muscle regeneration by remodeling the stem cell niche. Nature Communications, 12(1), 1-15.
- ↑ Rodríguez-Matellán, A., Alcazar, N., Hernández, F., Serrano, M., & Ávila, J. (2020). In Vivo Reprogramming Ameliorates Aging Features in Dentate Gyrus Cells and Improves Memory in Mice. Stem cell reports, 15(5), 1056-1066.
- ↑ Ocampo, A., Reddy, P., & Belmonte, J. C. I. (2016). Anti-aging strategies based on cellular reprogramming. Trends in molecular medicine, 22(8), 725-738.
- ↑ 19.0 19.1 Sinclair, D. A., & LaPlante, M. D. (2019). Lifespan: Why We Age—and Why We Don't Have To. Atria Books.
- ↑ Shannon, C. E. (1948). A mathematical theory of communication. The Bell system technical journal, 27(3), 379-423.
- ↑ Lepikhov, K., Arand, J., Wossidlo, M., & Walter, J. (2012). Epigenetic Reprogramming in Mammalian Development. In Encyclopedia of Molecular Cell Biology and Molecular Medicine. https://doi.org/10.1002/3527600906.mcb.201100038
- ↑ Kerepesi, C., Zhang, B., Lee, S.-G., Trapp, A., & Gladyshev, V. N. (2021). Epigenetic clocks reveal a rejuvenation event during embryogenesis followed by aging. Sci. Adv, 7, 6082–6107. https://www.science.org/doi/10.1126/sciadv.abg6082
- ↑ 23.0 23.1 Baylin, S. B., & Jones, P. A. (2016). Epigenetic determinants of cancer. Cold Spring Harbor Perspectives in Biology, 8(9). https://doi.org/10.1101/cshperspect.a019505
- ↑ Suvà, M. L., Riggi, N., & Bernstein, B. E. (2013). Epigenetic reprogramming in cancer. In Science (Vol. 340, Issue 6127, pp. 1567–1570). American Association for the Advancement of Science. https://doi.org/10.1126/science.1230184
- ↑ Wilson S, Filipp FV. A network of epigenomic and transcriptional cooperation encompassing an epigenomic master regulator in cancer. npj Systems Biology and Applications. 2018;4(1):24. doi:10.1038/s41540-018-0061-4
- ↑ Lu, Yuanjun, Chan, Y.-T., Tan, H.-Y., Li, S., Wang, N., & Feng, Y. (2020). Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Molecular Cancer 2020 19:1, 19(1), 1–16. https://doi.org/10.1186/S12943-020-01197-3
- ↑ 27.0 27.1 Zeps, N., Lysaght, T., Chadwick, R., Erler, A., Foo, R., Giordano, S., ... & Sugarman, J. (2021). Ethics and regulatory considerations for the clinical translation of somatic cell human epigenetic editing. Stem Cell Reports, 16(7), 1652-1655.
- ↑ Pagiatakis, C., Musolino, · Elettra, Gornati, R., Bernardini, G., & Papait, R. (2021). Epigenetics of aging and disease: a brief overview. Aging Clinical and Experimental Research, 33, 737–745. https://doi.org/10.1007/s40520-019-01430-0
- ↑ Noble, D. (2015). Conrad Waddington and the origin of epigenetics. The Journal of experimental biology, 218(6), 816-818.
- ↑ 30.0 30.1 30.2 30.3 de Lima Camillo, L. P., & Quinlan, R. B. (2021). A ride through the epigenetic landscape: aging reversal by reprogramming. GeroScience, 1-23.
- ↑ Ueda, J., Yamazaki, T., & Funakoshi, H. (2023). Toward the Development of Epigenome Editing-Based Therapeutics: Potentials and Challenges. International Journal of Molecular Sciences, 24(5), 4778. PMID: 36902207 PMC: 10003136 DOI: 10.3390/ijms24054778
- ↑ 32.0 32.1 32.2 Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. cell, 126(4), 663-676.
- ↑ 33.0 33.1 33.2 Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., & Yamanaka, S. (2007). Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell, 131(5), 861–872. https://doi.org/10.1016/j.cell.2007.11.019
- ↑ 34.0 34.1 Ocampo, A., Reddy, P., Martinez-Redondo, P., Platero-Luengo, A., Hatanaka, F., Hishida, T., Li, M., Lam, D., Kurita, M., Beyret, E., Araoka, T., Vazquez-Ferrer, E., Donoso, D., Roman, J. L., Xu, J., Rodriguez Esteban, C., Nuñez, G., Nuñez Delicado, E., Campistol, J. M., … Izpisua Belmonte, J. C. (2016). In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell, 167(7), 1719-1733.e12. https://doi.org/10.1016/j.cell.2016.11.052
- ↑ Aguirre M, Escobar M, Forero Amézquita S, Cubillos D, Rincón C, Vanegas P, Tarazona MP, Atuesta Escobar S, Blanco JC, Celis LG. (2023). Application of the Yamanaka Transcription Factors Oct4, Sox2, Klf4, and c-Myc from the Laboratory to the Clinic. Genes. 14(9):1697. https://doi.org/10.3390/genes14091697
- ↑ Olova, N., Simpson, D. J., Marioni, R. E., & Chandra, T. (2019). Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell, 18(1). https://doi.org/10.1111/acel.12877
- ↑ Mueller, M., Hermann, P. C., Liebau, S., Weidgang, C., Seufferlein, T., Kleger, A., & Perkhofer, L. (2016). The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem cell research, 16(2), 349-357.
- ↑ 38.0 38.1 38.2 Lin, T., & Wu, S. (2015). Reprogramming with small molecules instead of exogenous transcription factors. Stem cells international, 2015.
- ↑ Kim, Y., Jeong, J., & Choi, D. (2020). Small-molecule-mediated reprogramming: a silver lining for regenerative medicine. Experimental & molecular medicine, 52(2), 213-226.
- ↑ Tang, Y., & Cheng, L. (2017). Cocktail of chemical compounds robustly promoting cell reprogramming protects liver against acute injury. Protein & cell, 8(4), 273.
- ↑ Yang, J. H., Petty, C. A., Dixon-McDougall, T., Lopez, M. V., Tyshkovskiy, A., Maybury-Lewis, S., ... & Sinclair, D. A. (2023). Chemically induced reprogramming to reverse cellular aging. Aging (Albany NY), 15(13), 5966. PMID: 37437248 PMCID: PMC10373966 DOI: 10.18632/aging.204896
- ↑ Wang, J., Sun, S., & Deng, H. (2023). Chemical reprogramming for cell fate manipulation: Methods, applications, and perspectives. Cell Stem Cell. PMID: 37625410 DOI: 10.1016/j.stem.2023.08.001
- ↑ Fan, D., Liu, X., Shen, Z., Wu, P., Zhong, L., & Lin, F. (2023). Cell signaling pathways based on vitamin C and their application in cancer therapy. Biomedicine & Pharmacotherapy, 162, 114695. PMID: 37058822 DOI: 10.1016/j.biopha.2023.114695
- ↑ Young, J. I., Züchner, S., & Wang, G. (2015). Regulation of the epigenome by vitamin C. Annual review of nutrition, 35, 545-564. PMID: 25974700 PMCID: PMC4506708 DOI: 10.1146/annurev-nutr-071714-034228
- ↑ Liu, X., Khan, A., Li, H., Wang, S., Chen, X., & Huang, H. (2022). Ascorbic acid in epigenetic reprogramming. Current Stem Cell Research & Therapy, 17(1), 13-25. PMID: 34264189 DOI: 10.2174/1574888X16666210714152730
- ↑ 46.0 46.1 Product — turn.bio. (n.d.-b). Retrieved September 8, 2021, from https://www.turn.bio/product#pipeline-intro
- ↑ 47.0 47.1 Basu, A., & Tiwari, V. K. (2021). Epigenetic reprogramming of cell identity: lessons from development for regenerative medicine. Clinical Epigenetics, 13(1), 1-11.
- ↑ Understanding and Controlling in Vivo Reprogramming for Rejuvenation | Dr Manuel Serrano - YouTube. (n.d.). Retrieved October 11, 2021, from https://www.youtube.com/watch?v=X1q7C9MruLg
- ↑ Wilson, K. A., Chamoli, M., Hilsabeck, T. A., Pandey, M., Bansal, S., Chawla, G., & Kapahi, P. (2021). Evaluating the beneficial effects of dietary restrictions: A framework for precision nutrigeroscience. Cell Metabolism.
- ↑ 50.0 50.1 Shift Bioscience. (n.d.). Retrieved September 21, 2021, from https://www.shiftbioscience.com/
- ↑ Segal, D. J. (2020). Grand Challenges in Gene and Epigenetic Editing for Neurologic Disease. Frontiers in Genome Editing, 1, 1.
- ↑ 52.0 52.1 Lapasset, L., Milhavet, O., Prieur, A., Besnard, E., Babled, A., Aït-Hamou, N., ... & Lemaitre, J. M. (2011). Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes & development, 25(21), 2248-2253.
- ↑ Grigorash B.B, van Essen D, Liang G, Grosse L, Emelyanov A, Kang Z, Korablev A, Kanzler B, Molina C, Lopez E, Demidov O.N, Garrido C, Liu F, Saccani S, Bulavin D.V. (2023). p16High senescence restricts cellular plasticity during somatic cell reprogramming. Nat Cell Biol. PMID 37652981 doi:10.1038/s41556-023-01214-9
- ↑ Banito, A., Rashid, S. T., Acosta, J. C., Li, S., Pereira, C. F., Geti, I., ... & Gil, J. (2009). Senescence impairs successful reprogramming to pluripotent stem cells. Genes & development, 23(18), 2134-2139.
- ↑ 55.0 55.1 Mosteiro, L., Pantoja, C., Alcazar, N., Marión, R. M., Chondronasiou, D., Rovira, M., ... & Serrano, M. (2016). Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science, 354(6315).
- ↑ Chiche, A., Le Roux, I., von Joest, M., Sakai, H., Aguín, S. B., Cazin, C., ... & Li, H. (2017). Injury-induced senescence enables in vivo reprogramming in skeletal muscle. Cell stem cell, 20(3), 407-414.
- ↑ Chiche, A., Chen, C., & Li, H. (2020). The crosstalk between cellular reprogramming and senescence in aging and regeneration. Experimental gerontology, 138, 111005.
- ↑ Simon, A. K., Hollander, G. A., & McMichael, A. (2015). Evolution of the immune system in humans from infancy to old age. Proceedings of the Royal Society B: Biological Sciences, 282(1821), 20143085. PMID: 26702035 PMCID: PMC4707740 DOI: 10.1098/rspb.2014.3085
- ↑ Melendez, E., Chondronasiou, D., Mosteiro, L., Martínez de Villarreal, J., Fernández-Alfara, M., Lynch, C. J., … & Serrano, M. (2022). Natural killer cells act as an extrinsic barrier for in vivo reprogramming. Development, 149(8), dev200361. PMID 35420133 doi:10.1242/dev.200361
- ↑ AgeX Therapeutics | Technology. (n.d.). Retrieved September 21, 2021, from https://www.agexinc.com/technology/#iTR
- ↑ https://www.technologyreview.com/2021/09/04/1034364/altos-labs-silicon-valleys-jeff-bezos-milner-bet-living-forever/
- ↑ Meet Altos Labs, Silicon Valley’s latest wild bet on living forever | MIT Technology Review. (n.d.). Retrieved September 21, 2021, from https://www.technologyreview.com/2021/09/04/1034364/altos-labs-silicon-valleys-jeff-bezos-milner-bet-living-forever/
- ↑ Davis, M. (2001). Cancer institute director's exit leaves NIH in the lurch. Nature, 413(6853), 241-242.
- ↑ https://emea.illumina.com/company/news-center/press-releases/2013/1856858.html
- ↑ Cell identity reprogramming restores youthful gene expression | Calico research. (n.d.). Retrieved September 21, 2021, from https://reprog.research.calicolabs.com/
- ↑ Targeting the Biology of Aging - Epigenetic Reprogramming. (n.d.). Retrieved October 27, 2021, from https://www.lifebiosciences.com/our-science
Further reading
- Huyghe, A., Trajkova, A., & Lavial, F. (2023). Cellular plasticity in reprogramming, rejuvenation and tumorigenesis: a pioneer TF perspective. Trends in Cell Biology. https://doi.org/10.1016/j.tcb.2023.07.013
- Singh, P. B., & Zhakupova, A. (2022). Age reprogramming: cell rejuvenation by partial reprogramming. Development, 149(22). PMID: 36383700 PMC9845736 DOI: 10.1242/dev.200755
- Partial cellular reprogramming: A deep dive into an emerging rejuvenation technology. https://doi.org/10.1111/acel.14039
- Gene therapy mediated partial reprogramming extends lifespan and reverses age-related changes in aged mice. Preprint at bioRxiv, https://doi.org/10.1101/2023.01.04.522507 (2023)
- Liuyang, S., Wang, G., Wang, Y., He, H., Lyu, Y., Cheng, L., ... & Deng, H. (2023). Highly efficient and rapid generation of human pluripotent stem cells by chemical reprogramming. Cell Stem Cell, 30(4), 450-459. https://doi.org/10.1016/j.stem.2023.02.008
- Chemical reprogramming for cell fate manipulation: Methods, applications, and perspectives https://doi.org/10.1016/j.stem.2023.08.001
- Transplantation of Chemical Compound-Induced Cells from Human Fibroblasts Improves Locomotor Recovery in a Spinal Cord Injury Rat Model. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10530737/
- Mechanisms, pathways and strategies for rejuvenation through epigenetic reprogramming.
(December 2023) https://doi.org/10.1038/s43587-023-00539-2