Small ncRNAs influencing ageing and lifespan
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. Non-coding RNAs are endogenous transcripts that govern gene regulatory networks, thus impacting both physiological and pathological events. Non-coding RNAs constitute the majority of endogenous transcripts in the cells since human genome consists of only 3% protein-coding genes and most of the genome is transcribed to produce non-coding RNAs. ncRNA comprise numerous RNA species grouped in different classes, based on their different lengths and activities. Among these molecules, microRNAs, long non-coding RNAs, and more recently circular RNAs are considered crucial mediators of almost all cellular processes.[1][2]
MicroRNAs
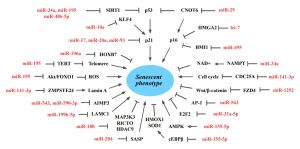
MicroRNAs (miRNAs) are endogenous small (∼22–25 nucleotides long) noncoding RNAs that control the expression of target mRNA by translational repression or mRNA degradation.[4] They are involved in many biological processes such as developmental timing, differentiation, cell death, stem cell proliferation and differentiation, immune response, aging and cancer. That's why miRNAs are the best characterized small non-coding RNAs influencing ageing and lifespan. Multiple miRNAs, including miRNA-1, miRNA-21, miRNA-22, miRNA-34a, miRNA-17, miRNA-145, miRNA-140, miRNA-106b, and miRNA-449a, are widely considered as critical regulators for cell senescence.[5][6][7][8][9][10]
Let-7, one of the first miRNAs discovered, was initially shown to control developmental timing in Caenorhabditis elegans[11] In mice, 12 genes encode members of the Let-7 family, which includes nine slightly different miRNAs [Let-7a, Let-c, Let-7f (all encoded by two genes), and Let-7b, Let-7d, Let-7e, Let-7g, Let-7i, and miR-98 (all encoded by one gene)]. Processing of Let-7 can be inhibited by an RNA-binding protein LIN28, that is highly expressed in embryonic stem cells.[12] Interestingly, Lin-28 can be used to achieve epigenetic reprogramming of human somatic cells into iPSC, even from centenarians fibroblasts.[13] Ectopic global LIN28a overexpression in mice was found to result in increased body size and crown–rump length, as well as increased glucose metabolism and insulin sensitivity.[14] LIN28A expression ends after development in most of tissues, and its re-expression in adult transgenic mice has been reported to enhance the regeneration of various somatic tissues by acting on somatic stem cells harbored within those tissues.[15][16] Knockout of Let-7 with a locked nucleic acid (LNA)-modified antimiR, that could inhibit Let-7 function in the whole body of mice, can reverse the glucose tolerance of diet-induced obese mice.[17]
Preliminary evidence suggests that beneficial effects of metformin may be due to regulation of let-7 expression, since "metformin no longer has potent antidiabetic actions in a liver-specific let-7 loss-of-function mouse model".[18][19]
MicroRNA-141-3p
Emerging evidence shows that the microRNA-141-3p is involved in various age-related pathologies. The mitochondria-related miR-141-3p might promote the pro-inflammatory cytokine (IL-6) expression, inducing the inflammatory response and contributing to the development of obesity. miR-141-3p over-expression reduced the tumor suppressor gene PTEN expression and promoted ATP production, oxidative stress, and the reduction of antioxidant capacity.[24] As a regulator of PPARγ (Peroxisome proliferator- activated receptor gamma), miR-143a-3p play an important role in adipogenesis via regulating MAPK7 (Mitogen-activated protein kinase 7) and fatty acid.[25]
Inhibiting miR-141-3p for three months with twice-weekly subcutaneous injections of Anti-miR-141-3p treatment improves musculoskeletal health with improving bone microstructure and muscle fiber size in aged mice. Molecular analysis revealed that miR-141-3p regulates the expression of AU-rich RNA-binding factor 1 (AUF1) and promotes the expression of the known muscle wasting transcription factor FOXO-1 (Forkhead transcription factor 1). It also promotes senescence (p21, p16) and pro-inflammatory (TNF-α, IL-1β, IFN-γ) environment whereas inhibiting miR-141-3p prevents these effects.[26]
miR-214-3p
miR-214-3p was downregulated in aged adipose stem cells (ASC)s, and its overexpression rejuvenated aged adipose stem cell (ASC).[27] A long non-coding RNA named double homeobox A pseudogene 10 (DUXAP10) located in the cytoplasm and functioned as a decoy for miR-214-3p is significantly accumulated in aged ASCs.[27] Knocking down DUXAP10 promoted stem cell proliferation and migration and halted cell senescence and the secretion of proinflammatory cytokines. Ras Association Domain Family Member 5 (RASSF5) was the target of miR-214-3p and was upregulated in aged ASCs. Overexpressing DUXAP10 and inhibiting miR-214-3p both enhanced RASSF5 content in ASCs, while DUXAP10 knockdown promoted the therapeutic ability of aged ASCs for skin wound healing.[27]
miRNA-34a
miR-34a has been implicated in cardiovascular fibrosis, dysfunction and related cardiovascular disorders as an essential regulator. There is a pivotal link among miR-34a, cardiovascular fibrosis, and Smad4/TGF-β1 signaling. miR-34a plays the critical roles in cardiovascular apoptosis, autophagy, inflammation, senescence and remodeling by modulating multifunctional signaling pathways.[28] MiR-34a accelerated the progression of atherosclerosis by regulating FOXO3 expression. It was reported that FOXO3 plays a critical role in restraining oxidative damage in ox-LDL-induced endothelial cell injury via the miR-34a/SIRT1/FOXO3 signaling pathway.[29][30] The expression of miR-34a increased in senescent MSCs cell culture with continuous passage.[31][32] MiR34a significantly reduced SIRT1 activity, NAD+ content, and NAD+/NADH ratio by targeting nicotinamide phosphoribosyl-transferase (NAMPT).[33]
The expression of miR-34a was strongly correlated with HbA1c level, suggesting that increased miR-34a expression is related to high glucose.[34][35]
miR-130b-5p
Patients with coronary artery disease (CAD) have high levels of miR-130b-5p in peripheral blood that correlated with severity of coronary artery disease.[37] Mechanistic studies revealed that miR-130b-5p mainly promoted the cardiomyocyte proliferation through the MAPK-ERK signaling pathway, and the dual-specific phosphatase 6 (Dusp6), a negative regulator of the MAPK-ERK signaling, was the direct target of miR-130b-5p. Moreover, overexpression of miR-130b-5p could promote the proliferation of cardiomyocytes and improve cardiac function in mice after myocardial infarction (MI).[38]
Human miR-130b-5p exhibited an impact upon the mRNA levels of a negative modulator of aging, Sprr1a when expressed in human primary dermal fibroblasts (HDFs), and induced cellular senescence.[39]
miR-200c-3p
Some studies have shown that the expression levels of miR-31-5p, miR-141-3p, and miR-200c-3p are elevated with age.[40] It was discovered that miR-200c-3p promoted the proliferation of adipose-derived MSCs and delayed cellular senescence.[41] miR-200c-3p can target and negatively regulate stearoyl-CoA desaturase 2 (SCD2) by binding to the 3′-UTR of SCD2 mRNA to restrain lipid synthesis in MSCs and thus reverse the inhibitory effect of SCD2 over-expression on MSC senescence.[42]
Small nuclear RNAs
Piwi-interacting RNA
References
- ↑ Zhang, P., Wu, W., Chen, Q., & Chen, M. (2019). Non-coding RNAs and their integrated networks. Journal of integrative bioinformatics, 16(3). PMID: 31301674 PMCID: PMC6798851 DOI: 10.1515/jib-2019-0027
- ↑ Varghese, L. N., Schwenke, D. O., & Katare, R. (2023). Role of noncoding RNAs in cardiac ageing. Frontiers in Cardiovascular Medicine, 10. PMID: 37034355 PMCID: PMC10073704 DOI: 10.3389/fcvm.2023.1142575
- ↑ He, L., Li, M., Liu, Z., Padhiar, A. A., & Zhou, G. (2023). Senescence of mesenchymal stem cells: implications in extracellular vesicles, miRNAs and their functional and therapeutic potentials. Aging Pathobiology and Therapeutics, 03-17. DOI: 10.31491/APT.2023.03.107
- ↑ Bushati, N., & Cohen, S. M. (2007). microRNA functions. Annu. Rev. Cell Dev. Biol., 23, 175-205. PMID: 17506695 DOI: 10.1146/annurev.cellbio.23.090506.123406
- ↑ Kinser, H. E., & Pincus, Z. (2020). MicroRNAs as modulators of longevity and the aging process. Human genetics, 139(3), 291-308. PMID: 31297598 PMCID: PMC6954352 DOI: 10.1007/s00439-019-02046-0
- ↑ Zia, A., Farkhondeh, T., Sahebdel, F., Pourbagher-Shahri, A. M., & Samarghandian, S. (2022). Key miRNAs in Modulating Aging and Longevity: A Focus on Signaling Pathways and Cellular Targets. Current Molecular Pharmacology, 15(5), 736-762. PMID: 34533452 DOI: 10.2174/1874467214666210917141541
- ↑ Ma, X., Zheng, Q., Zhao, G., Yuan, W., & Liu, W. (2020). Regulation of cellular senescence by microRNAs. Mechanisms of Ageing and Development, 189, 111264. PMID: 32450085 DOI: 10.1016/j.mad.2020.111264
- ↑ Azizidoost, S., Nasrolahi, A., Sheykhi-Sabzehpoush, M., Akiash, N., Assareh, A. R., Anbiyaee, O., ... & Kempisty, B. (2023). Potential roles of endothelial cells-related non-coding RNAs in cardiovascular diseases. Pathology-Research and Practice, 154330. https://doi.org/10.1016/j.prp.2023.154330
- ↑ Ortiz, G. G. R., Mohammadi, Y., Nazari, A., Ataeinaeini, M., Kazemi, P., Yasamineh, S., ... & Gholizadeh, O. (2023). A state-of-the-art review on the MicroRNAs roles in hematopoietic stem cell aging and longevity. Cell Communication and Signaling, 21(1), 1-16. PMID: 37095512 PMCID: PMC10123996 DOI: 10.1186/s12964-023-01117-0
- ↑ Chen, Z., Li, C., Huang, H., Shi, Y. L., & Wang, X. (2023). Research progress of aging-related microRNAs. Current Stem Cell Research & Therapy. PMID: 36892029 DOI: 10.2174/1574888X18666230308111043
- ↑ Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., ... & Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. nature, 403(6772), 901-906. PMID: 10706289 DOI: 10.1038/35002607
- ↑ Newman, M. A., & Hammond, S. M. (2010). Lin-28: an early embryonic sentinel that blocks Let-7 biogenesis. The international journal of biochemistry & cell biology, 42(8), 1330-1333. PMID: 20619222 DOI: 10.1016/j.biocel.2009.02.023
- ↑ Lapasset, L., Milhavet, O., Prieur, A., Besnard, E., Babled, A., Aït-Hamou, N., ... & Lemaitre, J. M. (2011). Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes & development, 25(21), 2248-2253. PMID: 22056670 PMCID: PMC3219229 DOI: 10.1101/gad.173922.111
- ↑ Zhu, H., Shah, S., Shyh-Chang, N., Shinoda, G., Einhorn, W. S., Viswanathan, S. R., ... & Daley, G. Q. (2010). Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature genetics, 42(7), 626-630. PMID: 20512147 PMCID: PMC3069638 DOI: 10.1038/ng.593
- ↑ Shyh-Chang, N., Zhu, H., De Soysa, T. Y., Shinoda, G., Seligson, M. T., Tsanov, K. M., ... & Daley, G. Q. (2013). Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell, 155(4), 778-792. PMID: 24209617 PMCID: PMC3917449 DOI: 10.1016/j.cell.2013.09.059
- ↑ Pieknell, K., Sulistio, Y. A., Wulansari, N., Darsono, W. H. W., Chang, M. Y., Ko, J. Y., ... & Lee, S. H. (2022). LIN28A enhances regenerative capacity of human somatic tissue stem cells via metabolic and mitochondrial reprogramming. Cell Death & Differentiation, 29(3), 540-555. PMID: 34556809 PMCID: PMC8901931 DOI: 10.1038/s41418-021-00873-1
- ↑ Frost, R. J., & Olson, E. N. (2011). Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences, 108(52), 21075-21080. PMID: 22160727 PMCID: PMC3248488 DOI: 10.1073/pnas.1118922109
- ↑ Xie, D., Chen, F., Zhang, Y., Shi, B., Song, J., Chaudhari, K., ... & Huang, Y. (2022). Let-7 underlies metformin-induced inhibition of hepatic glucose production. Proceedings of the National Academy of Sciences, 119(14), e2122217119. PMID: 35344434 PMCID: PMC9169108 DOI: 10.1073/pnas.2122217119
- ↑ Zhu, H., Jia, Z., Li, Y. R., & Danelisen, I. (2023). Molecular mechanisms of action of metformin: latest advances and therapeutic implications. Clinical and Experimental Medicine, 1-11. PMID: 37016064 PMCID: PMC10072049 DOI: 10.1007/s10238-023-01051-y
- ↑ Frost, R. J., & Olson, E. N. (2011). Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences, 108(52), 21075-21080. PMID: 22160727 PMCID: PMC3248488 DOI: 10.1073/pnas.1118922109
- ↑ Thornton, J. E., & Gregory, R. I. (2012). How does Lin28 let-7 control development and disease?. Trends in cell biology, 22(9), 474-482. PMID: 22784697 PMCID: PMC3432650 DOI: 10.1016/j.tcb.2012.06.001
- ↑ Wang, Y., Zhao, J., Chen, S., Li, D., Yang, J., Zhao, X., ... & Xu, L. (2022). Let-7 as a promising target in aging and aging-related diseases: a promise or a pledge. Biomolecules, 12(8), 1070. PMID: 36008964 PMCID: PMC9406090 DOI: 10.3390/biom12081070
- ↑ Cappelletti, C., Galbardi, B., Bruttini, M., Salerno, F., Canioni, E., Pasanisi, M. B., ... & Mantegazza, R. (2019). Aging‐associated genes and let‐7 microRNAs: a contribution to myogenic program dysregulation in oculopharyngeal muscular dystrophy. The FASEB Journal, 33(6), 7155-7167. PMID: 30860873 DOI: 10.1096/fj.201801577RR
- ↑ Ji, J., Qin, Y., Ren, J., Lu, C., Wang, R., Dai, X., ... & Wang, X. (2015). Mitochondria-related miR-141-3p contributes to mitochondrial dysfunction in HFD-induced obesity by inhibiting PTEN. Scientific reports, 5(1), 1-12. PMID: 26548909 PMCID: PMC4637860 DOI: 10.1038/srep16262
- ↑ Zhang, P., Du, J., Wang, L., Niu, L., Zhao, Y., Tang, G., ... & Zhu, L. (2018). MicroRNA-143a-3p modulates preadipocyte proliferation and differentiation by targeting MAPK7. Biomedicine & Pharmacotherapy, 108, 531-539. PMID: 30243086 DOI: 10.1016/j.biopha.2018.09.080
- ↑ Sagar Vyavahare , Sandeep Kumar , Kathryn Smith , Bharati Mendhe , Roger Zhong , Marion A. Cooley , Babak Baban , Carlos M. Isales , Mark Hamrick , William D Hill , Sadanand Fulzele. (2023). Inhibiting MicroRNA-141-3p Improves Musculoskeletal Health in Aged Mice. Aging and disease. 2023 https://doi.org/10.14336/AD.2023.0310-1
- ↑ 27.0 27.1 27.2 Ren, S., Li, C., Xiong, H., Wu, Q., Wu, X., Xiong, Z., ... & Chen, J. (2024). The Rejuvenation and Functional Restoration of Aged Adipose Stem Cells by DUXAP10 Knockdown via the Regulation of the miR-214-3p/RASSF5 Axis. Stem Cells Translational Medicine, szae015.
- ↑ Hua, C. C., Liu, X. M., Liang, L. R., Wang, L. F., & Zhong, J. C. (2022). Targeting the microRNA-34a as a novel therapeutic strategy for cardiovascular diseases. Frontiers in Cardiovascular Medicine, 8, 2243. PMID: 35155600 PMCID: PMC8828972 DOI: 10.3389/fcvm.2021.784044
- ↑ Zhang, H., Zhao, Z., Pang, X., Yang, J., Yu, H., Zhang, Y., ... & Zhao, J. (2017). MiR-34a/sirtuin-1/foxo3a is involved in genistein protecting against ox-LDL-induced oxidative damage in HUVECs. Toxicology Letters, 277, 115-122. PMID: 28688900 DOI: 10.1016/j.toxlet.2017.07.216
- ↑ Raucci, A., Macrì, F., Castiglione, S., Badi, I., Vinci, M. C., & Zuccolo, E. (2021). MicroRNA-34a: the bad guy in age-related vascular diseases. Cellular and Molecular Life Sciences, 1-24. PMID: 34698884 PMCID: PMC8629897 DOI: 10.1007/s00018-021-03979-4
- ↑ Mokhberian, N., Bolandi, Z., Eftekhary, M., Hashemi, S. M., Jajarmi, V., Sharifi, K., & Ghanbarian, H. (2020). Inhibition of miR-34a reduces cellular senescence in human adipose tissue-derived mesenchymal stem cells through the activation of SIRT1. Life Sciences, 257, 118055. PMID: 32634429 DOI: 10.1016/j.lfs.2020.118055
- ↑ Badi, I., Mancinelli, L., Polizzotto, A., Ferri, D., Zeni, F., Burba, I., et al. (2018). miR34a Promotes Vascular Smooth Muscle Cell Calcification by Downregulating SIRT1 (Sirtuin 1) and Axl (AXL Receptor Tyrosine Kinase). Atvb 38 (9), 2079–2090. doi:10.1161/atvbaha.118.311298
- ↑ Pi, C., Ma, C., Wang, H., Sun, H., Yu, X., Gao, X., ... & He, X. (2021). MiR-34a suppression targets Nampt to ameliorate bone marrow mesenchymal stem cell senescence by regulating NAD+-Sirt1 pathway. Stem Cell Research & Therapy, 12(1), 271. PMID: 33957971 PMCID: PMC8101138 DOI: 10.1186/s13287-021-02339-0
- ↑ Boon, R. A., Iekushi, K., Lechner, S., Seeger, T., Fischer, A., Heydt, S., ... & Dimmeler, S. (2013). MicroRNA-34a regulates cardiac ageing and function. Nature, 495(7439), 107-110. PMID: 23426265 DOI: 10.1038/nature11919
- ↑ Fomison-Nurse, I., Saw, E. E. L., Gandhi, S., Munasinghe, P. E., Van Hout, I., Williams, M. J. A., ... & Katare, R. (2018). Diabetes induces the activation of pro-ageing miR-34a in the heart, but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death & Differentiation, 25(7), 1336-1349. PMID: 29302057 PMCID: PMC6030067 DOI: 10.1038/s41418-017-0047-6
- ↑ Raucci, A., Macrì, F., Castiglione, S., Badi, I., Vinci, M. C., & Zuccolo, E. (2021). MicroRNA-34a: the bad guy in age-related vascular diseases. Cellular and Molecular Life Sciences, 1-24. PMID: 34698884 PMCID: PMC8629897 DOI: 10.1007/s00018-021-03979-4
- ↑ Coban, N., Ozuynuk, A. S., Erkan, A. F., Guclu-Geyik, F., & Ekici, B. (2021). Levels of miR-130b-5p in peripheral blood are associated with severity of coronary artery disease. Molecular Biology Reports, 48, 7719-7732. PMID: 34689283 DOI: 10.1007/s11033-021-06780-5
- ↑ Feng, K., Wu, Y., Li, J., Sun, Q., Ye, Z., Li, X., ... & Kang, J. (2024). Critical Role of miR-130b-5p in Cardiomyocyte Proliferation and Cardiac Repair in Mice After Myocardial Infarction. Stem Cells, 42(1), 29-41. PMID: 37933895 DOI: 10.1093/stmcls/sxad080
- ↑ Hong, J. Y., Nam, H. J., Ji, H., Kim, Y. Y., Hyun, M., Park, H. J., ... & McCrea, P. D. (2023). Sprr1 and miR-130b contribute to the senescence-like phenotype in aging. bioRxiv, 2023-10. PMID: 37961492 PMC10634805 DOI: 10.1101/2023.10.25.563779
- ↑ Capri, M., Olivieri, F., Lanzarini, C., Remondini, D., Borelli, V., Lazzarini, R., ... & Grazi, G. L. (2017). Identification of miR‐31‐5p, miR‐141‐3p, miR‐200c‐3p, and GLT 1 as human liver aging markers sensitive to donor–recipient age‐mismatch in transplants. Aging cell, 16(2), 262-272. PMID: 27995756 PMC5334540 DOI: 10.1111/acel.12549
- ↑ Anastasiadou, E., Ceccarelli, S., Messina, E., Gerini, G., Megiorni, F., Pontecorvi, P., ... & Marchese, C. (2021). MiR-200c-3p maintains stemness and proliferative potential in adipose-derived stem cells by counteracting senescence mechanisms. PLoS One, 16(9), e0257070. PMID: 34534238 PMC8448302 DOI: 10.1371/journal.pone.0257070
- ↑ Yu, X., Zhang, C., Ma, Q., Gao, X., Sun, H., Sun, Y., ... & He, X. (2024). SCD2 Regulation Targeted by miR-200c-3p on Lipogenesis Alleviates Mesenchymal Stromal Cell Senescence. International Journal of Molecular Sciences, 25(15), 8538. PMID: 39126105 PMC11313047 DOI: 10.3390/ijms25158538