Biological aging is not regarded as a single process, but rather as a complex group of interconnected cellular and molecular mechanisms. It is also worth noting that within the field of aging research there is also disagreement as to the actual concept of aging.[1]
The ¨Hallmarks of Aging¨ describe the basic processes thought to underlie aging in different organisms. These hallmarks are thought to be fundamental mechanisms shared across multiple major age-related diseases such as cancer, diabetes, cardiovascular disorders and neurodegenerative diseases. In a now widely-cited 2013 research paper, nine tentative hallmarks of aging were proposed.[2]
It is important to note that these hallmarks only offer a phenotypic observation of features associated with aging, but may fail to provide a causal explanation of aging (see section ´Critical Review´).
A decade after the original review, the "Hallmarks of Aging" has been updated and 3 new hallmarks have been added: disabled macroautophagy, chronic inflammation and dysbiosis.[3]
Criteria for the Hallmarks of Aging
López-Otín et al.[2] proposed in 2013 nine candidate aging traits thought to contribute to the aging phenotype and determine lifespan. These candidates arise from previous findings in research literature, with special focus on mammalian aging but also including key insights from simpler model organisms such as C. elegans.[4][5]
The criteria that each hallmark should, ideally, fulfil in order to be considered as an aging hallmark are:
- The hallmark should not only appear in a given mutant, but should in fact be present in wild-type aging models.
- Increasing the incidence of a hallmark (or degree to which is present) should lead to accelerated aging and shorter lifespan.
- Decreasing the incidence of a hallmark should lead to a decreased rate of aging and longer lifespan.
The authors of the study acknowledge that all three criteria are not met in many hallmarks, specially regarding criterion #3. They argue that complex relationships and interconnectedness between all hallmarks impedes observing beneficial lifespan effects when modifying just one aspect of aging. Furthermore, the lack of knowledge regarding how to experimentally ameliorate some hallmarks adds difficulty into validating this criterion.
Categories of the Hallmarks of Aging
The proposed Hallmarks of Aging are grouped into three main categories according to their hierarchy.
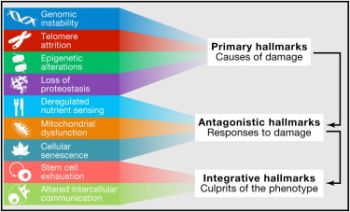
Primary hallmarks - defined as negative traits and sources of damage, which unequivocally lead to decreased health. This category includes:
- Genomic instability
- Telomere attrition
- Epigenetic alterations
- Loss of Proteostasiss
Antagonist hallmarks - defined as traits resulting from response to damage, and with varying effects on health depending on the intensity of their activity. For instance, low levels of cellular senescence might lead to health benefits, whilst high levels of cellular senesce might, in the opposite way, cause accelerated aging. This includes:
- Deregulated nutrient sensing
- Mitochondrial dysfunction
- Cellular senescence
Integrative hallmarks - defined as traits resulting from dysfunctional damage responses and considered to be the "culprits" of the aging phenotypes, given their direct negative effect on tissue homeostasis and function. This comprises:
- Stem cell exhaustion
- Altered intercellular communication
The 9 Hallmarks of Aging

1. Genomic instability
See the full article on DNA damage and repair
The genome is the total sum of DNA in our body and becomes damaged over time, with genomic instability leading to various age-related health problems.[2] DNA carries the information to make proteins that make up cells and tissues in living organisms.
DNA damage may be caused by UV radiation, X-ray radiation, chemical toxins such as tobacco.[8] The damage can also occur due to normal chemical reactions in our body, known as metabolism, and create byproducts (reactive oxygen species) that damage the DNA.[8]
As detailed in a review article, some scientists regard DNA damage as a unifying cause of the aging process, with causative interactions with all the so-called hallmarks of aging.[9]
The body has evolved repair systems to deal with the DNA damage as it arises and special proteins (enzymes) detect and repair broken strands of the DNA. However, the DNA repair processes are not perfect and errors in the DNA (mutations) arise over time. The body generally deals with these cells with too many mutations, through a kind of programmed cell suicide known as apoptosis. Alternatively, cells can undergo cellular senescence, a state of permanent cell cycle arrest that prevents further cell division. These senescent cells accumulate with aging and have been causatively linked to various age-related diseases and functional decline in mice.[10][11]
2. Telomere attrition
See the full article on telomeres.
Telomeres are regions of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. They protect the terminal regions of chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the ends of the DNA strand for a double strand break.
Telomere shortening is associated with aging, mortality and aging-related diseases.[2] Normal aging is associated with telomere shortening in both humans and mice, and studies on genetically modified animal models suggest causal links between telomere erosion and aging.[12] Leonard Hayflick demonstrated that a normal human fetal cell population will divide between 40 and 60 times in cell culture before entering a senescence phase.[13] Each time a cell undergoes mitosis, the telomeres on the ends of each chromosome shorten slightly. Cell division will cease once telomeres shorten to a critical length. This is useful when uncontrolled cell proliferation (like in cancer) needs to be stopped, but detrimental when normally functioning cells are unable to divide when necessary.
An enzyme called telomerase elongates telomeres in gametes (reproductive cells) and embryonic stem cells. Telomerase deficiency in humans has been linked to several aging-related diseases related to loss of regenerative capacity of tissues.[14] It has also been shown that premature aging in telomerase-deficient mice is reverted when telomerase is reactivated.[15]
3. Epigenetic alterations
Learn about epigenetic clocks.
Out of all the genes that make up a genome, only a subset are expressed at any given time. The functioning of a genome depends both on the specific order of its nucleotides (genomic factors), and also on which sections of the DNA chain are spooled tightly on histones and thus rendered inaccessible, and which ones are unspooled and available for transcription (epigenomic factors). Depending on the needs of the specific tissue type and environment that a given cell is in, histones can be modified to turn specific genes on or off as needed. The profile of where, when and to what extent these modifications occur (the epigenetic profile) changes with aging, turning useful genes off and unnecessary ones on, disrupting the normal functioning of the cell.
As an example, sirtuins are a type of protein deacetylase that promotes the binding of DNA onto histones and thus controls gene expression by turning them of or off. These enzymes use NAD as a cofactor. As we age, the level of NAD in our cells decreases and so does the ability of certain sirtuins to turn off unneeded genes at the right time. Decreasing the activity of certain sirtuins has been associated with accelerated aging and increasing their activity has been shown to stave off several age-related diseases.
4. Loss of proteostasis
See the full article on proteostasis.
Proteostasis is the homeostatic process of maintaining all the proteins necessary for the functioning of the cell in their proper shape, structure and abundance.[2] Protein misfolding, oxidation, abnormal cleavage or undesired post-translational modification can create dysfunctional or even toxic proteins or protein aggregates that hinder the normal functioning of the cell. Though these proteins are continually removed and recycled, formation of damaged or aggregated proteins increases with age, leading to a gradual loss of proteostasis. Based on animal studies, this can be slowed or suppressed by caloric restriction or by administration of rapamycin, which is partly mediated through inhibiting the mTOR pathway.
Loss of proteostasis has been linked to various age-related diseases. Neurodegenerative diseases are prominent examples of this, for example, Alzheimer's is associated with the accumulation of proteins that are regarded as toxic to brain neurons, such as amyloid beta and tau.[16][17] The role of aging in the loss of proteostasis has been shown to improve diseases such as Alzheimer's and Parkinson's, via investigating rapamycin or chaperone-mediated autophagy.[17][18][19]
5. Deregulated nutrient-sensing
See the full article on metabolic flexibility.
Nutrient sensing is a cell's ability to recognize, and respond to, changes in the concentration of macronutrients such as glucose, fatty acids and amino acids. In times of abundance, anabolism is induced through various pathways, the most well-studied among them the mTOR pathway. When energy and nutrients are scarce, proteins such as the AMPK receptor senses this and turns down mTOR to conserve resources.
In a growing organism, growth and cell proliferation are important and thus mTOR is upregulated. In a fully grown organism, mTOR-activating signals naturally decline during aging. It has been found that forcibly overactivating these pathways in grown mice leads to accelerated aging and increased incidence of cancer. mTOR inhibition methods like dietary restriction or rapamycin have been shown to be one of the most robust methods of increasing healthy lifespan in worms, flies and mice.
Metabolic flexibility is the ability of an organism to adapt efficiently and rapidly to different metabolic and energy states. This has shown to be dependent on functional mitochondrial dynamics.[20] Loss of metabolic flexibility (or the proper activation/suppression of the AMPK and mTOR pathways) during aging has been proposed as the main risk factor for the onset of age-related diseases.[20]
6. Mitochondrial dysfunction
See the full article on mitochondrial dysfunction.
The mitochondria is the powerhouse of the cell. Different human cells can contain up to thousands of mitochondria, each one converting carbon (in the form of acetyl-CoA) and oxygen into energy (in the form of ATP) and carbon dioxide during cellular respiration.
During aging, the efficiency of mitochondria tends to decrease. The reasons for this are still quite unclear, but several mechanisms are suspected - reduced biogenesis, accumulation of damage and mutations in mitochondrial DNA, oxidation of mitochondrial proteins, and defective quality control by mitophagy. Dysfunctional mitochondria contribute to aging through interfering with intercellular signalling and triggering inflammatory reactions.
Mitochondria are highly dynamic organelles that undergo coordinated cycles of fission and fusion in order to maintain their shape, distribution and size, as shown by imaging studies in live cells. Functional mitochondrial dynamics are key for regulating organismal homeostasis and are required to maintain appropriate metabolic flexibility.
7. Cellular senescence
See the full article on cellular senescence.
Under certain conditions, a cell will exit the cell cycle without dying, instead becoming dormant and ceasing its normal function. This is called cellular senescence. Senescence can be induced by several factors, including telomere shortening, DNA damage and stress. Since the immune system is programmed to seek out and eliminate senescent cells, it might be that senescence is one way for the body to rid itself of cells damaged beyond repair. Compounds targeting the removal of senescent cells are known as senolytics.
The links between cell senescence and aging are several:
- The proportion of senescent cells increases with age.
- Senescent cells secrete inflammatory markers which may contribute to aging.
- Clearance of senescent cells has been found to delay the onset of age-related disorders.
8. Stem cell exhaustion
Stem cells are undifferentiated or partially differentiated cells that can proliferate indefinitely. For the first few days after fertilization, the embryo consists almost entirely of stem cells. As the fetus grows, the cells multiply, differentiate and assume their appropriate function within the organism. In adults, stem cells are mostly located in areas that undergo gradual wear (intestine, lung, mucosa, skin) or need continuous replenishment (red blood cells, immune cells, sperm cells, hair follicles).
Loss of regenerative ability is one of the most obvious consequences of aging. This is largely because the proportion of stem cells and the speed of their division gradually lowers over time. It has been found that stem cell rejuvenation can reverse some of the effects of aging at the organismal level.
9. Altered intercellular communication
Different tissues and the cells they consist of need to orchestrate their work in a tightly controlled manner so that the organism as a whole can function. One of the main ways this is achieved is through excreting signal molecules into the blood where they make their way to other tissues, affecting their behavior. The profile of these molecules changes as we age.
One of the most prominent changes in cell signaling biomarkers is "inflammaging", the development of a chronic low-grade inflammation throughout the body with advanced age. The normal role of inflammation is to recruit the body's immune system and repair mechanisms to a specific damaged area for as long as the damage and threat are present. The constant presence of inflammation markers throughout the body wears out the immune system and damages healthy tissue.
It's also been found that senescent cells excrete a specific set of molecules called the SASP (Senescence-Associated Secretory Phenotype) which induce senescence in neighboring cells. Conversely, lifespan-extending manipulations targeting one tissue can slow the aging process in other tissues as well.
10. Cell size
[21] [22] [23] [24] [25] [26] [27]
Correlation between aging and nucleolar size
Critical Review
While the nine proposed Hallmarks of Aging are believed to each contribute to the aging process itself, definitive evidence is missing. As mentioned previously, many of the hallmarks fail to fulfil the three criteria by which they were selected for. Scientists are, therefore, not yet able to confirm whether ameliorating these hallmarks would indeed lead to a decelerated aging phenotype and increased lifespan, which would provide evidence that these hallmarks are upstream of the aging process.
Consequently, these hallmarks do not offer a causal explanation as to why we age, and could potentially just offer a phenotypic observation of associated aging features. In other words, these observed hallmarks might be a result of an upstream unknown aging process, similar to how the appearance of wrinkles or white hairs is known to be associated to older individuals.
Another important aspect of the Hallmarks of Aging by López-Otín et al., is the limitation of its scope to a whole-organism level. The latest high-throughout RNA-seq studies, including from the longevity-research giant Calico, have demonstrated that aging is largely regulated in a tissue-specific manner and that age-dependent changes vary enormously according to cell type.[28][29] In fact, down-regulation of energy metabolism pathways during aging is the only universal change observed across all cell types.[28] This highlights the importance of building a more comprehensive tissue-specific reference, for the hallmarks of aging, as it has already been explored in the context of neurodegenerative diseases.[30]
PanSci, a single-cell transcriptome atlas profiling over 20 million cells from 623 mouse tissue samples, encompassing a range of organs across different life stages, sexes, and genotypes allowed to identify more than 3,000 unique cellular states and catalog over 200 distinct aging-associated cell populations experiencing significant depletion or expansion. Panoramic analysis uncovered temporally structured, organ- and lineage-specific shifts of cellular dynamics during lifespan progression. Including aging-associated alterations in immune cell populations, revealing both widespread shifts and organ-specific changes.[31]
Nonetheless, and similarly to the case of epigenetic clocks, the Hallmarks of Aging might provide a useful framework to examine the effect of rejuvenating interventions, until more exhaustive studies of the hallmarks of aging are available at the tissue level.
Biomarkers of aging
References
- ↑ Cohen, A. A., Legault, V., & Fülöp, T. (2020). What if there’s no such thing as “aging”?. Mechanisms of Ageing and Development, 192, 111344.
- ↑ 2.0 2.1 2.2 2.3 2.4 López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., & Kroemer, G. (2013). The hallmarks of aging. Cell, 153(6), 1194-1217.
- ↑ López-Otín, C. et al. (2023) “Hallmarks of aging: An expanding universe,” Cell, 186(2), pp. 243–278. Available at: https://doi.org/10.1016/j.cell.2022.11.001.
- ↑ Kenyon, C. (2010). The genetics of ageing. Nature, 464(7288), 504-512. doi: 10.1038/nature08980
- ↑ Gems, D., & Partridge, L. (2013). Genetics of Longevity in Model Organisms: Debates and Paradigm Shifts. Annual Review Of Physiology, 75(1), 621-644. doi: 10.1146/annurev-physiol-030212-183712
- ↑ López-Otín, C., Blasco, M., Partridge, L., Serrano, M., & Kroemer, G. (2013). The Hallmarks of Aging. Cell, 153(6), 1194-1217. doi: 10.1016/j.cell.2013.05.039
- ↑ López-Otín, C., Blasco, M., Partridge, L., Serrano, M., & Kroemer, G. (2013). The Hallmarks of Aging. Cell, 153(6), 1194-1217. doi: 10.1016/j.cell.2013.05.039
- ↑ 8.0 8.1 Rastogi, R. P., Kumar, A., Tyagi, M. B., & Sinha, R. P. (2010). Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. Journal of nucleic acids, 2010.
- ↑ Schumacher, B., Pothof, J., Vijg, J., & Hoeijmakers, J. H. (2021). The central role of DNA damage in the ageing process. Nature, 592(7856), 695-703.
- ↑ Calcinotto, A., Kohli, J., Zagato, E., Pellegrini, L., Demaria, M., & Alimonti, A. (2019). Cellular senescence: aging, cancer, and injury. Physiological reviews, 99(2), 1047-1078.
- ↑ Baker, D. J., Wijshake, T., Tchkonia, T., LeBrasseur, N. K., Childs, B. G., Van De Sluis, B., ... & Van Deursen, J. M. (2011). Clearance of p16 Ink4a-positive senescent cells delays ageing-associated disorders. Nature, 479(7372), 232-236.
- ↑ Muñoz-Lorente, M. A., Cano-Martin, A. C., & Blasco, M. A. (2019). Mice with hyper-long telomeres show less metabolic aging and longer lifespans. Nature communications, 10(1), 1-14.
- ↑ Hayflick, L. (1965). The limited in vitro lifetime of human diploid cell strains. Experimental cell research, 37(3), 614-636.
- ↑ de Jesus, B. B., & Blasco, M. A. (2013). Telomerase at the intersection of cancer and aging. Trends in genetics, 29(9), 513-520.
- ↑ Jaskelioff, M., Muller, F. L., Paik, J. H., Thomas, E., Jiang, S., Adams, A. C., ... & DePinho, R. A. (2011). Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature, 469(7328), 102-106.
- ↑ Kaeberlein, M., & Galvan, V. (2019). Rapamycin and Alzheimer’s disease: time for a clinical trial?. Science translational medicine, 11(476).
- ↑ 17.0 17.1 Spilman, P., Podlutskaya, N., Hart, M. J., Debnath, J., Gorostiza, O., Bredesen, D., ... & Galvan, V. (2010). Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-β levels in a mouse model of Alzheimer's disease. PloS one, 5(4), e9979.
- ↑ Yao, R. Q., Qi, D. S., Yu, H. L., Liu, J., Yang, L. H., & Wu, X. X. (2012). Quercetin attenuates cell apoptosis in focal cerebral ischemia rat brain via activation of BDNF–TrkB–PI3K/Akt signaling pathway. Neurochemical research, 37(12), 2777-2786.
- ↑ Pupyshev, A. B., Tikhonova, M. A., Akopyan, A. A., Tenditnik, M. V., Dubrovina, N. I., & Korolenko, T. A. (2019). Therapeutic activation of autophagy by combined treatment with rapamycin and trehalose in a mouse MPTP-induced model of Parkinson's disease. Pharmacology Biochemistry and Behavior, 177, 1-11.
- ↑ 20.0 20.1 Weir, H., Yao, P., Huynh, F., Escoubas, C., Goncalves, R., & Burkewitz, K. et al. (2017). Dietary Restriction and AMPK Increase Lifespan via Mitochondrial Network and Peroxisome Remodeling. Cell Metabolism, 26(6), 884-896.e5. doi: 10.1016/j.cmet.2017.09.024
- ↑ Lengefeld, J., Cheng, C. W., Maretich, P., Blair, M., Hagen, H., McReynolds, M. R., ... & Amon, A. (2021). Cell size is a determinant of stem cell potential during aging. Science advances, 7(46), eabk0271.
- ↑ Biran, A., Zada, L., Abou Karam, P., Vadai, E., Roitman, L., Ovadya, Y., ... & Krizhanovsky, V. (2017). Quantitative identification of senescent cells in aging and disease. Aging cell, 16(4), 661-671. PMID: 28455874 PMC5506427 DOI: 10.1111/acel.12592
- ↑ Vidal, P. J., Pérez, A. P., Yahya, G., & Aldea, M. (2024). Transcriptomic balance and optimal growth are determined by cell size. Molecular Cell. https://doi.org/10.1016/j.molcel.2024.07.005
- ↑ Pérez-Ortín, J. E., García-Marcelo, M. J., Delgado-Román, I., Muñoz-Centeno, M. C., & Chávez, S. (2024). Influence of cell volume on the gene transcription rate. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 195008. PMID: 38246270 DOI: 10.1016/j.bbagrm.2024.195008
- ↑ Lanz, M. C., Zatulovskiy, E., Swaffer, M. P., Zhang, L., Ilerten, I., Zhang, S., ... & Skotheim, J. M. (2022). Increasing cell size remodels the proteome and promotes senescence. Molecular cell, 82(17), 3255-3269. PMID: 35987199 PMC9444988 DOI: 10.1016/j.molcel.2022.07.017
- ↑ Manohar, S., & Neurohr, G. E. (2024). Too big not to fail: emerging evidence for size‐induced senescence. The FEBS Journal, 291(11), 2291-2305. PMID: 37986656 DOI: 10.1111/febs.16983
- ↑ Khurana, A., Chadha, Y., & Schmoller, K. M. (2023). Too big not to fail: Different paths lead to senescence of enlarged cells. Molecular Cell, 83(22), 3946-3947. DOI:https://doi.org/10.1016/j.molcel.2023.10.024
- ↑ 28.0 28.1 Roux, A., Yuan, H., Podshivalova, K., Hendrickson, D., Kerr, R., Kenyon, C., & Kelley, D. (2022). The complete cell atlas of an aging multicellular organism. doi: 10.1101/2022.06.15.496201
- ↑ Wang, X., Jiang, Q., Song, Y., He, Z., Zhang, H., & Song, M. et al. (2022). Ageing induces tissue‐specific transcriptomic changes in Caenorhabditis elegans. The EMBO Journal, 41(8). doi: 10.15252/embj.2021109633
- ↑ Mattson, M., & Arumugam, T. (2018). Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States. Cell Metabolism, 27(6), 1176-1199. doi: 10.1016/j.cmet.2018.05.011
- ↑ Zhang, Z., Schaefer, C., Jiang, W., Lu, Z., Lee, J., Sziraki, A., ... & Cao, J. (2024). A Panoramic View of Cell Population Dynamics in Mammalian Aging. bioRxiv, 2024-03. https://doi.org/10.1101/2024.03.01.583001
- ↑ Aging Biomarker Consortium., Bao, H., Cao, J. et al. (2023). Biomarkers of aging. Sci. China Life Sci. https://doi.org/10.1007/s11427-023-2305-0